| (1) AGO.mut | (3) AGO1.ip | (11) AGO2.ip | (1) AGO3.ip | (14) B-CELL | (20) BRAIN | (1) CELL-LINE | (1) DCR.mut | (1) DGCR8.mut | (11) EMBRYO | (5) ESC | (5) FIBROBLAST | (1) KIDNEY | (6) LIVER | (5) LYMPH | (15) OTHER | (1) OTHER.mut | (1) PANCREAS | (5) SKIN | (8) SPLEEN | (7) TESTES | (2) THYMUS | (2) TOTAL-RNA | (1) UTERUS |
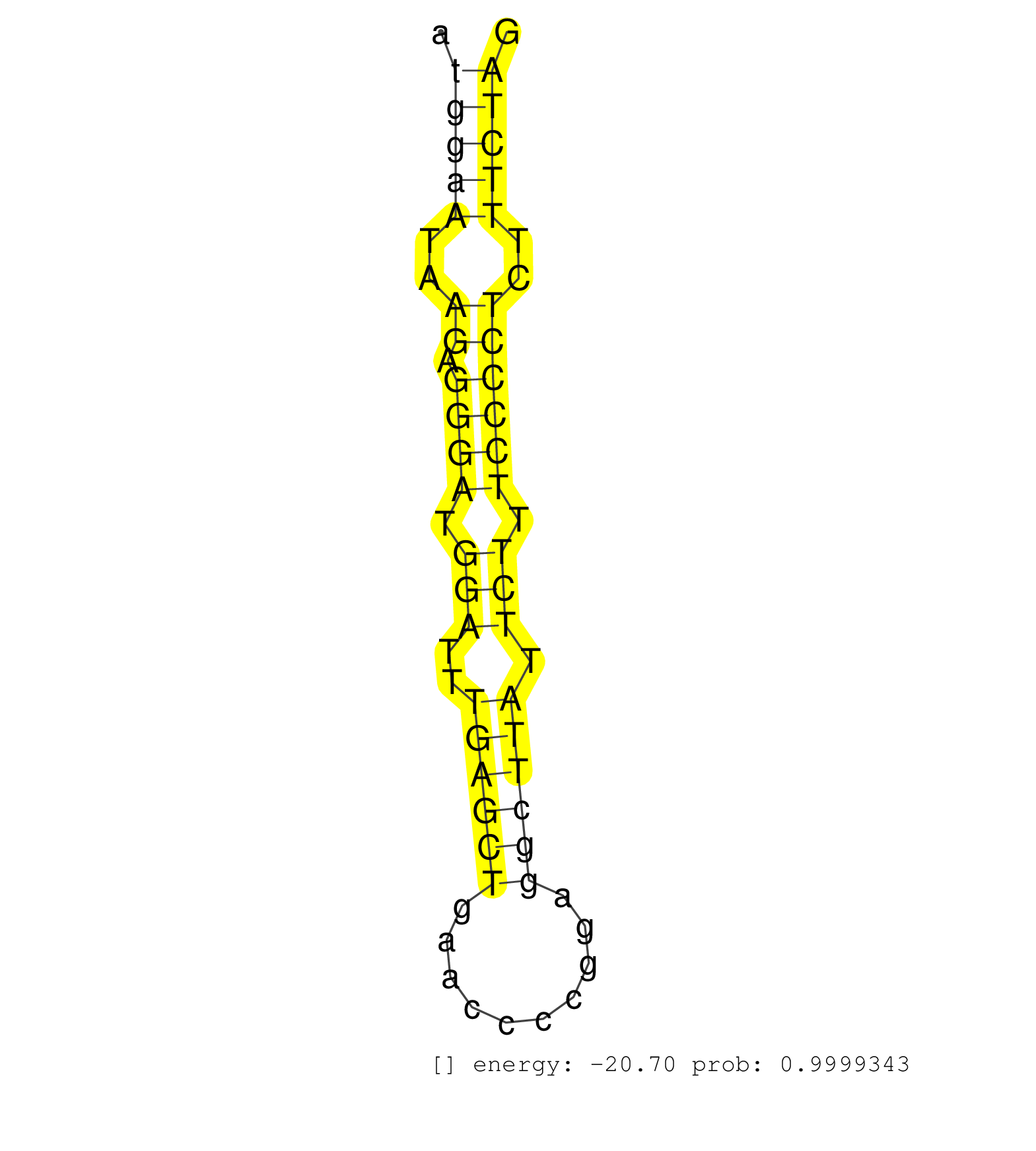
| GAGCAAGGCCGGGTAAATATTTAACTTGGTAGAGTCCCAGGTGAGTCTCTCATCCCTGAACCCCCATCTTCTCCTGTGTAGTCACCAAGAATGCCTGGTTATTCTTTGCCTTCGTTAGAAGGTGAGATGGTGTGAGACATGGAATAAGAGGGATGGATTTGAGCTGAACCCCGGAGGCTTATTCTTTCCCCTCTTTCTAGGCTGAACCATCCCAATGTGGTGGCTGCCCGGGATGTCCCAGAGGGGATGC ...........................................................................................................................................(((((..((.((((.(((..((((((..........)))))).))).))))))..)))))................................................... ..........................................................................................................................................139..........................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR042458(GSM539850) mouse basophil cells [09-002]. (b cell) | SRR042471(GSM539863) mouse TFHS cells [09-002]. (spleen) | SRR042446(GSM539838) mouse mature B cells (spleen) replicate 1 [09-002]. (b cell) | SRR042469(GSM539861) mouse Th2 cells [09-002]. (lymph) | SRR042463(GSM539855) mouse natural killer cells [09-002]. (spleen) | SRR059769(GSM562831) Treg_Drosha. (spleen) | SRR042452(GSM539844) mouse B1 B cells [09-002]. (b cell) | SRR306531(GSM750574) 19-24nt. (ago2 brain) | SRR042468(GSM539860) mouse Th1 cells [09-002]. (lymph) | SRR042459(GSM539851) mouse neutrophil cells replicate 1 [09-002]. (blood) | SRR042457(GSM539849) mouse mast cells [09-002]. (bone marrow) | SRR042447(GSM539839) mouse mature B cells (spleen) replicate 2 [09-002]. (b cell) | SRR042467(GSM539859) mouse T cells activated in vitro with Concanavalin A [09-002]. (spleen) | SRR345203(SRX097264) source: size fractionated RNA from mouse hipp. (brain) | SRR042473(GSM539865) mouse nTreg cells [09-002]. (lymph) | SRR042466(GSM539858) mouse CD8+ T cells (spleen) [09-002]. (spleen) | SRR042465(GSM539857) mouse CD4+ T cells (spleen) [09-002]. (spleen) | SRR042454(GSM539846) mouse germinal center B cells (lymph node) [09-002]. (b cell) | SRR306542(GSM750585) 19-24nt. (ago2 brain) | SRR042470(GSM539862) mouse Th17 cells [09-002]. (lymph) | SRR042487(GSM539879) mouse BclXL B activated with LPS/IL4 [BalbC] [09-002]. (spleen) | mjLiverWT1() Liver Data. (liver) | SRR042448(GSM539840) mouse B cells activated in vitro with LPS/IL4 replicate 1 [09-002]. (b cell) | SRR042445(GSM539837) mouse immature B cells [09-002]. (b cell) | SRR306529(GSM750572) 19-24nt. (ago2 brain) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR391847(GSM852142) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR042488(GSM539880) mouse BclXL germinal center B cells [BalbC] [09-002]. (B cell) | SRR042461(GSM539853) mouse dendritic cells [09-002]. (bone marrow) | SRR042472(GSM539864) mouse iTreg cells [09-002]. (lymph) | SRR073954(GSM629280) total RNA. (blood) | SRR042444(GSM539836) mouse pre B cells [09-002]. (b cell) | SRR390299(GSM849857) cell line: NIH-3T3cell type: fibroblastip ant. (ago2 fibroblast) | SRR345204(SRX097265) source: size fractionated RNA from mouse hipp. (brain) | SRR059766(GSM562828) DN3_Drosha. (thymus) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR546155(SRX180174) Global profiling of miRNA and the hairpin pre. (ago2 Neuroblastoma) | SRR042443(GSM539835) mouse pro B cells [09-002]. (b cell) | SRR065055(SRR065055) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | SRR042451(GSM539843) mouse B cells activated in vitro with LPS/aIgD-Dextran [09-002]. (b cell) | SRR306534(GSM750577) 19-24nt. (ago2 brain) | SRR042474(GSM539866) mouse embryonic stem cells [09-002]. (ESC) | SRR039185(GSM485325) shLuc.1309;ROSA-rtTA (-dox). (fibroblast) | SRR042464(GSM539856) mouse CD4+CD8+ T cells (thymus) [09-002]. (thymus) | SRR345205(SRX097266) source: size fractionated RNA from mouse hipp. (brain) | SRR042450(GSM539842) mouse B cells activated in vitro with LPS/IL4 replicate 3 [09-002]. (b cell) | SRR039186(GSM485326) shLuc.1309;ROSA-rtTA (+dox). (fibroblast) | SRR525244(SRA056111/SRX170320) Mus musculus domesticus miRNA sequencing. (ago2 Blood) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | SRR279903(GSM689053) cell type: mouse embryonic stem cellcell line. (ESC) | SRR042475(GSM539867) mouse embryonic fibroblast cells [09-002]. (fibroblast) | SRR037924(GSM510462) e9p5_rep2. (embryo) | SRR042456(GSM539848) mouse hematopoetic progenitor cells [09-002]. (bone marrow) | SRR042449(GSM539841) mouse B cells activated in vitro with LPS/IL4 replicate 2 [09-002]. (b cell) | SRR306536(GSM750579) 19-24nt. (ago2 brain) | SRR065053(SRR065053) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR060845(GSM561991) total RNA. (brain) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR306533(GSM750576) 19-24nt. (ago2 brain) | Ago2IP517(Rui) Quantitative functions of Argonaute proteins . (ago2 skin) | SRR553586(SRX182792) source: Testis. (testes) | Ago1IP812(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | SRR346414(SRX098255) Global profiling of miRNA and the hairpin pre. (Spleen) | SRR394084(GSM855970) background strain: C57BL6/SV129cell type: KRa. (dicer cell line) | SRR553583(SRX182789) source: Cerebellum. (Cerebellum) | SRR065048(SRR065048) Tissue-specific Regulation of Mouse MicroRNA . (liver) | SRR042479(GSM539871) mouse liver tissue [09-002]. (liver) | GSM510452(GSM510452) newborn_rep8. (total RNA) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR206942(GSM723283) other. (brain) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (ago2 liver) | SRR042480(GSM539872) mouse kidney tissue [09-002]. (kidney) | SRR037928(GSM510466) e7p5_rep2. (embryo) | SRR525240(SRA056111/SRX170316) Mus musculus domesticus miRNA sequencing. (ago1 Blood) | SRR037927(GSM510465) e7p5_rep1. (embryo) | SRR345201(SRX097262) source: size fractionated RNA from mouse hipp. (brain) | SRR039154(GSM471931) HL1_siSrf2_smallRNAseq. (muscle) | SRR037925(GSM510463) e9p5_rep3. (embryo) | SRR038741(GSM527276) small RNA-Seq. (brain) | SRR042453(GSM539845) mouse marginal zone B cells (spleen) [09-002]. (b cell) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR065056(SRR065056) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | GSM314552(GSM314552) ESC wild type (Illumina). (ESC) | Ago3IP812(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR042482(GSM539874) mouse skin tissue [09-002]. (skin) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM640579(GSM640579) small RNA in the liver with paternal Low pro. (liver) | SRR039153(GSM471930) HL1_siSrf1_smallRNAseq. (muscle) | mjTestesWT1() Testes Data. (testes) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR039184(GSM485324) shLuc.1309 (+dox). (fibroblast) | Ago1IP517(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | GSM509276(GSM509276) small RNA cloning by length. (testes) | SRR032476(GSM485234) sRNA_delayed_deep_sequencing. (uterus) | SRR345206(SRX097267) source: size fractionated RNA from mouse hipp. (brain) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR402766(SRX117949) source: ES PGK female cells. (ESC) | SRR065058(SRR065058) Tissue-specific Regulation of Mouse MicroRNA . (pancreas) | SRR345202(SRX097263) source: size fractionated RNA from mouse hipp. (brain) | SRR042477(GSM539869) mouse brain tissue [09-002]. (brain) | SRR525238(SRA056111/SRX170314) Second IgG control. (Blood) | SRR037914(GSM510451) newborn_rep7. (total RNA) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | GSM475281(GSM475281) total RNA. (testes) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR279904(GSM689054) cell type: mouse embryonic stem cellcell line. (ESC) | SRR346417(SRX098258) Global profiling of miRNA and the hairpin pre. (Brain) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................ATAAGAGGGATGGATTTGAGCT..................................................................................... | 22 | 1 | 49.00 | 49.00 | 6.00 | 1.00 | 2.00 | 1.00 | 4.00 | 2.00 | 2.00 | - | - | 1.00 | - | - | 1.00 | 1.00 | - | - | 1.00 | 1.00 | - | 2.00 | 1.00 | - | - | - | - | 1.00 | - | 1.00 | 1.00 | 1.00 | 1.00 | 2.00 | - | 3.00 | 1.00 | 1.00 | - | - | - | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - |
| ..............................................................................................................................................AATAAGAGGGATGGATTTGAGCT..................................................................................... | 23 | 1 | 42.00 | 42.00 | 1.00 | - | 2.00 | - | 2.00 | 1.00 | 1.00 | 2.00 | - | - | - | - | 3.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | 2.00 | 1.00 | - | 1.00 | - | - | 1.00 | - | - | 1.00 | 3.00 | - | - | - | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | 1.00 | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | 1.00 | 1.00 | - | 1.00 | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................AATAAGAGGGATGGATTTGAGC...................................................................................... | 22 | 1 | 27.00 | 27.00 | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | 1.00 | 1.00 | - | 2.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..............................................................................................................................................AATAAGAGGGATGGATTTGAG....................................................................................... | 21 | 1 | 22.00 | 22.00 | 8.00 | 2.00 | - | 1.00 | - | - | - | - | - | 1.00 | 1.00 | 1.00 | - | - | - | 2.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................ATAAGAGGGATGGATTTGAG....................................................................................... | 20 | 1 | 19.00 | 19.00 | 4.00 | - | - | 1.00 | - | - | 1.00 | - | - | 2.00 | - | 1.00 | - | - | 1.00 | - | 2.00 | - | - | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................ATAAGAGGGATGGATTTGAGC...................................................................................... | 21 | 1 | 11.00 | 11.00 | - | 1.00 | - | - | - | - | - | 1.00 | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................AATAAGAGGGATGGATTT.......................................................................................... | 18 | 1 | 10.00 | 10.00 | 3.00 | 3.00 | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................AATAAGAGGGATGGATTTGA........................................................................................ | 20 | 1 | 10.00 | 10.00 | 1.00 | - | - | 1.00 | - | 3.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..............................................................................................................................................AATAAGAGGGATGGATTTG......................................................................................... | 19 | 1 | 9.00 | 9.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................ATAAGAGGGATGGATTTGAGCTT.................................................................................... | 23 | 1 | 8.00 | 49.00 | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................TAAGAGGGATGGATTTGAGCT..................................................................................... | 21 | 1 | 7.00 | 7.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................TAAGAGGGATGGATTTGAG....................................................................................... | 19 | 1 | 7.00 | 7.00 | - | 1.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................ATAAGAGGGATGGATTTGA........................................................................................ | 19 | 1 | 7.00 | 7.00 | 1.00 | 2.00 | - | - | - | - | - | - | 2.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................ATAAGAGGGATGGATTTG......................................................................................... | 18 | 1 | 4.00 | 4.00 | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................AATAAGAGGGATGGATTTGAGCA..................................................................................... | 23 | 1 | 4.00 | 27.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TTATTCTTTCCCCTCTTTCTAGT................................................. | 23 | 3.00 | 0.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................TTGGTAGAGTCCCAGATT............................................................................................................................................................................................................... | 18 | 3.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................TTGGTAGAGTCCCAGATTG.............................................................................................................................................................................................................. | 19 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................TAAGAGGGATGGATTTGCGCT..................................................................................... | 21 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................ATAAGAGGGATGGATTTGAGA...................................................................................... | 21 | 1 | 2.00 | 19.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................AATAAGAGGGATGGATTTGAGA...................................................................................... | 22 | 1 | 2.00 | 22.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................TAAGAGGGATGGATTTGA........................................................................................ | 18 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................ATAAGAGGGATGGATTTGAGCTAA................................................................................... | 24 | 1 | 2.00 | 49.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................ATAAGAGGGATGGATTTGAGCAA.................................................................................... | 23 | 1 | 1.00 | 11.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................ATAAGAGGGATGGATTTGAGT...................................................................................... | 21 | 1 | 1.00 | 19.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................TAAGAGGGATGGATTTGAGCTTAT.................................................................................. | 24 | 1 | 1.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................AATAAGAGGGATGGATTTGAGAAA.................................................................................... | 24 | 1 | 1.00 | 22.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...............................................................................................................................................ATAAGAGGGATGGATTTGAGCTAAA.................................................................................. | 25 | 1 | 1.00 | 49.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................TAAGAGGGATGGATTTGAGCTTA................................................................................... | 23 | 1 | 1.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................TAAGAGGGATGGATTTGAGCTGA................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................ACATGGAATAAGAGGGATGGATT........................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...............................................................................................................................................ATAAGAGGGATGGATTTGAGCTAC................................................................................... | 24 | 1 | 1.00 | 49.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................CAATGTGGTGGCTGCCCGGGATGT.............. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................TGGCTGCCCGGGATGTCCCAGAGG...... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................TAAGAGGGATGGATTTGAGC...................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................GAGCTGAACCCCGGAGCT........................................................................ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................ATAAGAGGGATGGATTTGCGCT..................................................................................... | 22 | 1 | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................AATAAGAGGGATGGATTTTA........................................................................................ | 20 | 1 | 1.00 | 10.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................GAATGCCTGGTTATTCT................................................................................................................................................. | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................GAATAAGAGGGATGGATTTGAGC...................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................TAAGAGGGATGGATTTGAGCTGAAC................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................GGTGGCTGCCCGGGAT................ | 16 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ................................................................................................................................................TAAGAGGGATGGATTTGAGCA..................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................ATAAGAGGGATGGATTTGAGCA..................................................................................... | 22 | 1 | 1.00 | 11.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................AATAAGAGGGATGGATTTGCGC...................................................................................... | 22 | 1 | 1.00 | 9.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................AATAAGAGGGATGGATTTGAAA...................................................................................... | 22 | 1 | 1.00 | 10.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................ATGGAATAAGAGGGAGG............................................................................................... | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................AGGGATGGATTTGAGGTCC................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................GAATAAGAGGGATGGATTTGA........................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TTATTCTTTCCCCTCTTTCT.................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..............................................................................................................................................AATAAGAGGGATGGATTTGTGCT..................................................................................... | 23 | 1 | 1.00 | 9.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................ATAAGAGGGATGGATTTGAGCTA.................................................................................... | 23 | 1 | 1.00 | 49.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TCGTTAGAAGGTGA............................................................................................................................. | 14 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................TGGATTTGAGCTGAAC................................................................................. | 16 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 |
| GAGCAAGGCCGGGTAAATATTTAACTTGGTAGAGTCCCAGGTGAGTCTCTCATCCCTGAACCCCCATCTTCTCCTGTGTAGTCACCAAGAATGCCTGGTTATTCTTTGCCTTCGTTAGAAGGTGAGATGGTGTGAGACATGGAATAAGAGGGATGGATTTGAGCTGAACCCCGGAGGCTTATTCTTTCCCCTCTTTCTAGGCTGAACCATCCCAATGTGGTGGCTGCCCGGGATGTCCCAGAGGGGATGC ...........................................................................................................................................(((((..((.((((.(((..((((((..........)))))).))).))))))..)))))................................................... ..........................................................................................................................................139..........................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR042458(GSM539850) mouse basophil cells [09-002]. (b cell) | SRR042471(GSM539863) mouse TFHS cells [09-002]. (spleen) | SRR042446(GSM539838) mouse mature B cells (spleen) replicate 1 [09-002]. (b cell) | SRR042469(GSM539861) mouse Th2 cells [09-002]. (lymph) | SRR042463(GSM539855) mouse natural killer cells [09-002]. (spleen) | SRR059769(GSM562831) Treg_Drosha. (spleen) | SRR042452(GSM539844) mouse B1 B cells [09-002]. (b cell) | SRR306531(GSM750574) 19-24nt. (ago2 brain) | SRR042468(GSM539860) mouse Th1 cells [09-002]. (lymph) | SRR042459(GSM539851) mouse neutrophil cells replicate 1 [09-002]. (blood) | SRR042457(GSM539849) mouse mast cells [09-002]. (bone marrow) | SRR042447(GSM539839) mouse mature B cells (spleen) replicate 2 [09-002]. (b cell) | SRR042467(GSM539859) mouse T cells activated in vitro with Concanavalin A [09-002]. (spleen) | SRR345203(SRX097264) source: size fractionated RNA from mouse hipp. (brain) | SRR042473(GSM539865) mouse nTreg cells [09-002]. (lymph) | SRR042466(GSM539858) mouse CD8+ T cells (spleen) [09-002]. (spleen) | SRR042465(GSM539857) mouse CD4+ T cells (spleen) [09-002]. (spleen) | SRR042454(GSM539846) mouse germinal center B cells (lymph node) [09-002]. (b cell) | SRR306542(GSM750585) 19-24nt. (ago2 brain) | SRR042470(GSM539862) mouse Th17 cells [09-002]. (lymph) | SRR042487(GSM539879) mouse BclXL B activated with LPS/IL4 [BalbC] [09-002]. (spleen) | mjLiverWT1() Liver Data. (liver) | SRR042448(GSM539840) mouse B cells activated in vitro with LPS/IL4 replicate 1 [09-002]. (b cell) | SRR042445(GSM539837) mouse immature B cells [09-002]. (b cell) | SRR306529(GSM750572) 19-24nt. (ago2 brain) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR391847(GSM852142) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR042488(GSM539880) mouse BclXL germinal center B cells [BalbC] [09-002]. (B cell) | SRR042461(GSM539853) mouse dendritic cells [09-002]. (bone marrow) | SRR042472(GSM539864) mouse iTreg cells [09-002]. (lymph) | SRR073954(GSM629280) total RNA. (blood) | SRR042444(GSM539836) mouse pre B cells [09-002]. (b cell) | SRR390299(GSM849857) cell line: NIH-3T3cell type: fibroblastip ant. (ago2 fibroblast) | SRR345204(SRX097265) source: size fractionated RNA from mouse hipp. (brain) | SRR059766(GSM562828) DN3_Drosha. (thymus) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR546155(SRX180174) Global profiling of miRNA and the hairpin pre. (ago2 Neuroblastoma) | SRR042443(GSM539835) mouse pro B cells [09-002]. (b cell) | SRR065055(SRR065055) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | SRR042451(GSM539843) mouse B cells activated in vitro with LPS/aIgD-Dextran [09-002]. (b cell) | SRR306534(GSM750577) 19-24nt. (ago2 brain) | SRR042474(GSM539866) mouse embryonic stem cells [09-002]. (ESC) | SRR039185(GSM485325) shLuc.1309;ROSA-rtTA (-dox). (fibroblast) | SRR042464(GSM539856) mouse CD4+CD8+ T cells (thymus) [09-002]. (thymus) | SRR345205(SRX097266) source: size fractionated RNA from mouse hipp. (brain) | SRR042450(GSM539842) mouse B cells activated in vitro with LPS/IL4 replicate 3 [09-002]. (b cell) | SRR039186(GSM485326) shLuc.1309;ROSA-rtTA (+dox). (fibroblast) | SRR525244(SRA056111/SRX170320) Mus musculus domesticus miRNA sequencing. (ago2 Blood) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | SRR279903(GSM689053) cell type: mouse embryonic stem cellcell line. (ESC) | SRR042475(GSM539867) mouse embryonic fibroblast cells [09-002]. (fibroblast) | SRR037924(GSM510462) e9p5_rep2. (embryo) | SRR042456(GSM539848) mouse hematopoetic progenitor cells [09-002]. (bone marrow) | SRR042449(GSM539841) mouse B cells activated in vitro with LPS/IL4 replicate 2 [09-002]. (b cell) | SRR306536(GSM750579) 19-24nt. (ago2 brain) | SRR065053(SRR065053) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR060845(GSM561991) total RNA. (brain) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR306533(GSM750576) 19-24nt. (ago2 brain) | Ago2IP517(Rui) Quantitative functions of Argonaute proteins . (ago2 skin) | SRR553586(SRX182792) source: Testis. (testes) | Ago1IP812(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | SRR346414(SRX098255) Global profiling of miRNA and the hairpin pre. (Spleen) | SRR394084(GSM855970) background strain: C57BL6/SV129cell type: KRa. (dicer cell line) | SRR553583(SRX182789) source: Cerebellum. (Cerebellum) | SRR065048(SRR065048) Tissue-specific Regulation of Mouse MicroRNA . (liver) | SRR042479(GSM539871) mouse liver tissue [09-002]. (liver) | GSM510452(GSM510452) newborn_rep8. (total RNA) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR206942(GSM723283) other. (brain) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (ago2 liver) | SRR042480(GSM539872) mouse kidney tissue [09-002]. (kidney) | SRR037928(GSM510466) e7p5_rep2. (embryo) | SRR525240(SRA056111/SRX170316) Mus musculus domesticus miRNA sequencing. (ago1 Blood) | SRR037927(GSM510465) e7p5_rep1. (embryo) | SRR345201(SRX097262) source: size fractionated RNA from mouse hipp. (brain) | SRR039154(GSM471931) HL1_siSrf2_smallRNAseq. (muscle) | SRR037925(GSM510463) e9p5_rep3. (embryo) | SRR038741(GSM527276) small RNA-Seq. (brain) | SRR042453(GSM539845) mouse marginal zone B cells (spleen) [09-002]. (b cell) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR065056(SRR065056) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | GSM314552(GSM314552) ESC wild type (Illumina). (ESC) | Ago3IP812(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR042482(GSM539874) mouse skin tissue [09-002]. (skin) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM640579(GSM640579) small RNA in the liver with paternal Low pro. (liver) | SRR039153(GSM471930) HL1_siSrf1_smallRNAseq. (muscle) | mjTestesWT1() Testes Data. (testes) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR039184(GSM485324) shLuc.1309 (+dox). (fibroblast) | Ago1IP517(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | GSM509276(GSM509276) small RNA cloning by length. (testes) | SRR032476(GSM485234) sRNA_delayed_deep_sequencing. (uterus) | SRR345206(SRX097267) source: size fractionated RNA from mouse hipp. (brain) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR402766(SRX117949) source: ES PGK female cells. (ESC) | SRR065058(SRR065058) Tissue-specific Regulation of Mouse MicroRNA . (pancreas) | SRR345202(SRX097263) source: size fractionated RNA from mouse hipp. (brain) | SRR042477(GSM539869) mouse brain tissue [09-002]. (brain) | SRR525238(SRA056111/SRX170314) Second IgG control. (Blood) | SRR037914(GSM510451) newborn_rep7. (total RNA) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | GSM475281(GSM475281) total RNA. (testes) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR279904(GSM689054) cell type: mouse embryonic stem cellcell line. (ESC) | SRR346417(SRX098258) Global profiling of miRNA and the hairpin pre. (Brain) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................TGAGCTGAACCCCGGCA.......................................................................... | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................TTATTCTTTCCCCTCTTT...................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |