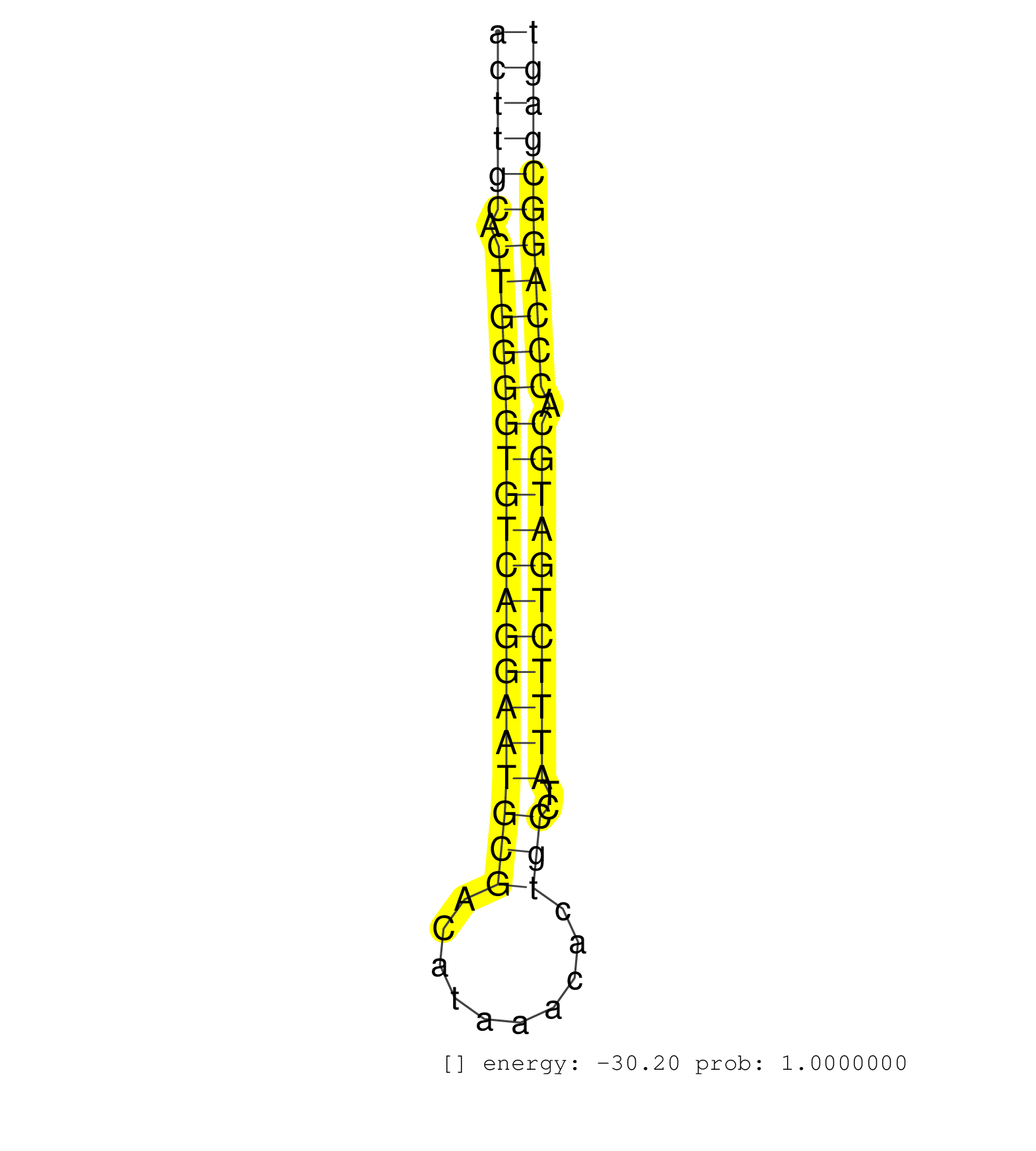
| (1) AGO1.ip | (5) AGO2.ip | (1) AGO3.ip | (16) BRAIN | (1) CELL-LINE | (1) DCR.mut | (1) DGCR8.mut | (5) EMBRYO | (2) FIBROBLAST | (2) LIVER | (2) LYMPH | (5) OTHER | (1) OTHER.mut | (3) SKIN | (5) SPLEEN | (6) TESTES | (1) THYMUS |
| TCTGTCACTTAGCAACAGTTAAAGCCCAGGCATCTGTATCATGTCAGATCACCTGTGTTGGTATTGCCTTCTGCCATATCAAATTAGCTGGTTTTACATGCAAGCTGGAAGCATAATGCCAAGCAGAGACTGTGTGCGAGCCACTTGCACTGGGGTGTCAGGAATGCGACATAAACACTGCCTATTTCTGATGCACCCAGGCGAGTGGCAGTGTGATGAGCGTTTACAGTGGAGACTTTGGCAACCTAGA ..............................................................................................................................................((((((.(((((((((((((((((((..........)))..))))))))))).)))))))))))............................................ ..............................................................................................................................................143............................................................206.......................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR059769(GSM562831) Treg_Drosha. (spleen) | SRR553582(SRX182788) source: Brain. (Brain) | SRR042463(GSM539855) mouse natural killer cells [09-002]. (spleen) | SRR206941(GSM723282) other. (brain) | SRR553583(SRX182789) source: Cerebellum. (Cerebellum) | SRR553602(SRX182808) source: Sertoli cells. (Sertoli cells) | SRR346417(SRX098258) Global profiling of miRNA and the hairpin pre. (Brain) | SRR306532(GSM750575) 19-24nt. (ago2 brain) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR059770(GSM562832) Treg_Dicer. (spleen) | SRR038744(GSM527279) small RNA-Seq. (brain) | SRR042473(GSM539865) mouse nTreg cells [09-002]. (lymph) | SRR346423(SRX098265) Global profiling of miRNA and the hairpin pre. (Brain) | SRR042465(GSM539857) mouse CD4+ T cells (spleen) [09-002]. (spleen) | Ago2IP504(Rui) Quantitative functions of Argonaute proteins . (ago2 skin) | SRR059765(GSM562827) DN3_control. (thymus) | SRR346414(SRX098255) Global profiling of miRNA and the hairpin pre. (Spleen) | GSM640579(GSM640579) small RNA in the liver with paternal Low pro. (liver) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | Ago1IP812(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR042483(GSM539875) mouse muscle tissue [09-002]. (muscle) | SRR039610(GSM527274) small RNA-Seq. (brain) | SRR346418(SRX098259) Global profiling of miRNA and the hairpin pre. (Liver) | SRR345207(SRX097268) source: size fractionated RNA from mouse hipp. (brain) | SRR306540(GSM750583) 19-24nt. (ago2 brain) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR038743(GSM527278) small RNA-Seq. (dicer brain) | SRR039184(GSM485324) shLuc.1309 (+dox). (fibroblast) | SRR206940(GSM723281) other. (brain) | SRR306526(GSM750569) 19-24nt. (ago2 brain) | SRR073954(GSM629280) total RNA. (blood) | SRR345202(SRX097263) source: size fractionated RNA from mouse hipp. (brain) | SRR306527(GSM750570) 19-24nt. (ago2 brain) | Ago3IP805(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | SRR345205(SRX097266) source: size fractionated RNA from mouse hipp. (brain) | SRR685339(GSM1079783) Small RNAs (15-50 nts in length) from immorta. (cell line) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR042469(GSM539861) mouse Th2 cells [09-002]. (lymph) | SRR346421(SRX098263) Global profiling of miRNA and the hairpin pre. (mixture) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR037900(GSM510436) testes_rep1. (testes) | SRR039186(GSM485326) shLuc.1309;ROSA-rtTA (+dox). (fibroblast) | GSM475281(GSM475281) total RNA. (testes) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................CACTGGGGTGTCAGGAATGCGAC................................................................................ | 23 | 1 | 9.00 | 9.00 | - | - | 1.00 | 2.00 | 1.00 | - | - | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................CACTGGGGTGTCAGGAATGCG.................................................................................. | 21 | 1 | 8.00 | 8.00 | - | 1.00 | - | 1.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................CACTGGGGTGTCAGGAATGCGA................................................................................. | 22 | 1 | 6.00 | 6.00 | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...................................................................................................................................................CACTGGGGTGTCAGGAATGCGACA............................................................................... | 24 | 1 | 5.00 | 5.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................ACTGGGGTGTCAGGAATGCGA................................................................................. | 21 | 1 | 5.00 | 5.00 | 1.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................TGGGGTGTCAGGAATGCGACA............................................................................... | 21 | 1 | 3.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ....................................................................................................................................................................................CCTATTTCTGATGCACCCAGTT................................................ | 22 | 3.00 | 0.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................CCTATTTCTGATGCACCCAGACT............................................... | 23 | 2.00 | 0.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................CCTATTTCTGATGCACCCAGAT................................................ | 22 | 2.00 | 0.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................CCTATTTCTGATGCACCCAGA................................................. | 21 | 2.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................CCTATTTCTGATGCACCCAGTCT............................................... | 23 | 2.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................CCTATTTCTGATGCACCCAGT................................................. | 21 | 2.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................CACTGGGGTGTCAGGAATGC................................................................................... | 20 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................CTGGGGTGTCAGGAATGCGAC................................................................................ | 21 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ....................................................................................................................................................................................CCTATTTCTGATGCACCCAGGA................................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................TGAGCGTTTACAGTGGAGACTTT........... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................ACTGGGGTGTCAGGAATGCGAC................................................................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................AGTGGCAGTGTGATGAGCGTT.......................... | 21 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................CACTGGGGTGTCAGGAATGCGACT............................................................................... | 24 | 1 | 1.00 | 9.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................TTAAAGCCCAGGCATCTAA..................................................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | |
| ...................TAAAGCCCAGGCATCTAATT................................................................................................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | |
| ....................................................................................................................................................................................CCTATTTCTGATGCACCC.................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................CCTATTTCTGATGCACCCAGACC............................................... | 23 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................TGGGGTGTCAGGAATGCGACAT.............................................................................. | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................GGGGTGTCAGGAATGCGA................................................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................TGCACTGGGGTGTCAGGAATGCGACATA............................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................CACTGGGGTGTCAGGAATGCGT................................................................................. | 22 | 1 | 1.00 | 8.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................ACTGGGGTGTCAGGAATGCGACA............................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................CCTATTTCTGATGCACCCAGTCA............................................... | 23 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................CCTATTTCTGATGCACCCATTT................................................ | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................................CTGGGGTGTCAGGAATGCGA................................................................................. | 20 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................CAGAGACTGTGTGCGCCC............................................................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................CCTATTTCTGATGCACCCAGG................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................AGAGACTGTGTGCGAGCCACTTGCACTGGGGTGTCAGGAATGCGACATAAACACTGCCTATTTCTGATGCACCCAG.................................................. | 76 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ....................................................................................................................................................................................................................GTGATGAGCGTTTACAGTGGAGACTTTG.......... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................GTGTGATGAGCGTTTACAGTGGAGACTTT........... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................TGCACTGGGGTGTCAGGAATGC................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................CCTATTTCTGATGCACCCAGAAA............................................... | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................ACTGGGGTGTCAGGAATGCG.................................................................................. | 20 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................AGGCATCTGTATCATGCTAA........................................................................................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................CACTGGGGTGTCAGGAATGCTA................................................................................. | 22 | 1 | 1.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................CCTATTTCTGATGCACCCAGTC................................................ | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................CCAGGCATCTGTATC.................................................................................................................................................................................................................. | 15 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 |
| TCTGTCACTTAGCAACAGTTAAAGCCCAGGCATCTGTATCATGTCAGATCACCTGTGTTGGTATTGCCTTCTGCCATATCAAATTAGCTGGTTTTACATGCAAGCTGGAAGCATAATGCCAAGCAGAGACTGTGTGCGAGCCACTTGCACTGGGGTGTCAGGAATGCGACATAAACACTGCCTATTTCTGATGCACCCAGGCGAGTGGCAGTGTGATGAGCGTTTACAGTGGAGACTTTGGCAACCTAGA ..............................................................................................................................................((((((.(((((((((((((((((((..........)))..))))))))))).)))))))))))............................................ ..............................................................................................................................................143............................................................206.......................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR059769(GSM562831) Treg_Drosha. (spleen) | SRR553582(SRX182788) source: Brain. (Brain) | SRR042463(GSM539855) mouse natural killer cells [09-002]. (spleen) | SRR206941(GSM723282) other. (brain) | SRR553583(SRX182789) source: Cerebellum. (Cerebellum) | SRR553602(SRX182808) source: Sertoli cells. (Sertoli cells) | SRR346417(SRX098258) Global profiling of miRNA and the hairpin pre. (Brain) | SRR306532(GSM750575) 19-24nt. (ago2 brain) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR059770(GSM562832) Treg_Dicer. (spleen) | SRR038744(GSM527279) small RNA-Seq. (brain) | SRR042473(GSM539865) mouse nTreg cells [09-002]. (lymph) | SRR346423(SRX098265) Global profiling of miRNA and the hairpin pre. (Brain) | SRR042465(GSM539857) mouse CD4+ T cells (spleen) [09-002]. (spleen) | Ago2IP504(Rui) Quantitative functions of Argonaute proteins . (ago2 skin) | SRR059765(GSM562827) DN3_control. (thymus) | SRR346414(SRX098255) Global profiling of miRNA and the hairpin pre. (Spleen) | GSM640579(GSM640579) small RNA in the liver with paternal Low pro. (liver) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | Ago1IP812(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | GSM509275(GSM509275) MitoPLD+/+ E16.5 small RNA. (testes) | SRR042483(GSM539875) mouse muscle tissue [09-002]. (muscle) | SRR039610(GSM527274) small RNA-Seq. (brain) | SRR346418(SRX098259) Global profiling of miRNA and the hairpin pre. (Liver) | SRR345207(SRX097268) source: size fractionated RNA from mouse hipp. (brain) | SRR306540(GSM750583) 19-24nt. (ago2 brain) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR038743(GSM527278) small RNA-Seq. (dicer brain) | SRR039184(GSM485324) shLuc.1309 (+dox). (fibroblast) | SRR206940(GSM723281) other. (brain) | SRR306526(GSM750569) 19-24nt. (ago2 brain) | SRR073954(GSM629280) total RNA. (blood) | SRR345202(SRX097263) source: size fractionated RNA from mouse hipp. (brain) | SRR306527(GSM750570) 19-24nt. (ago2 brain) | Ago3IP805(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | SRR345205(SRX097266) source: size fractionated RNA from mouse hipp. (brain) | SRR685339(GSM1079783) Small RNAs (15-50 nts in length) from immorta. (cell line) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR042469(GSM539861) mouse Th2 cells [09-002]. (lymph) | SRR346421(SRX098263) Global profiling of miRNA and the hairpin pre. (mixture) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR037900(GSM510436) testes_rep1. (testes) | SRR039186(GSM485326) shLuc.1309;ROSA-rtTA (+dox). (fibroblast) | GSM475281(GSM475281) total RNA. (testes) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................................................................CTGATGCACCCAGGCTGCA............................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................................ATAAACACTGCCTATAGA.............................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................GCCTATTTCTGATGCACCCAGGCGAGTGG.......................................... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |