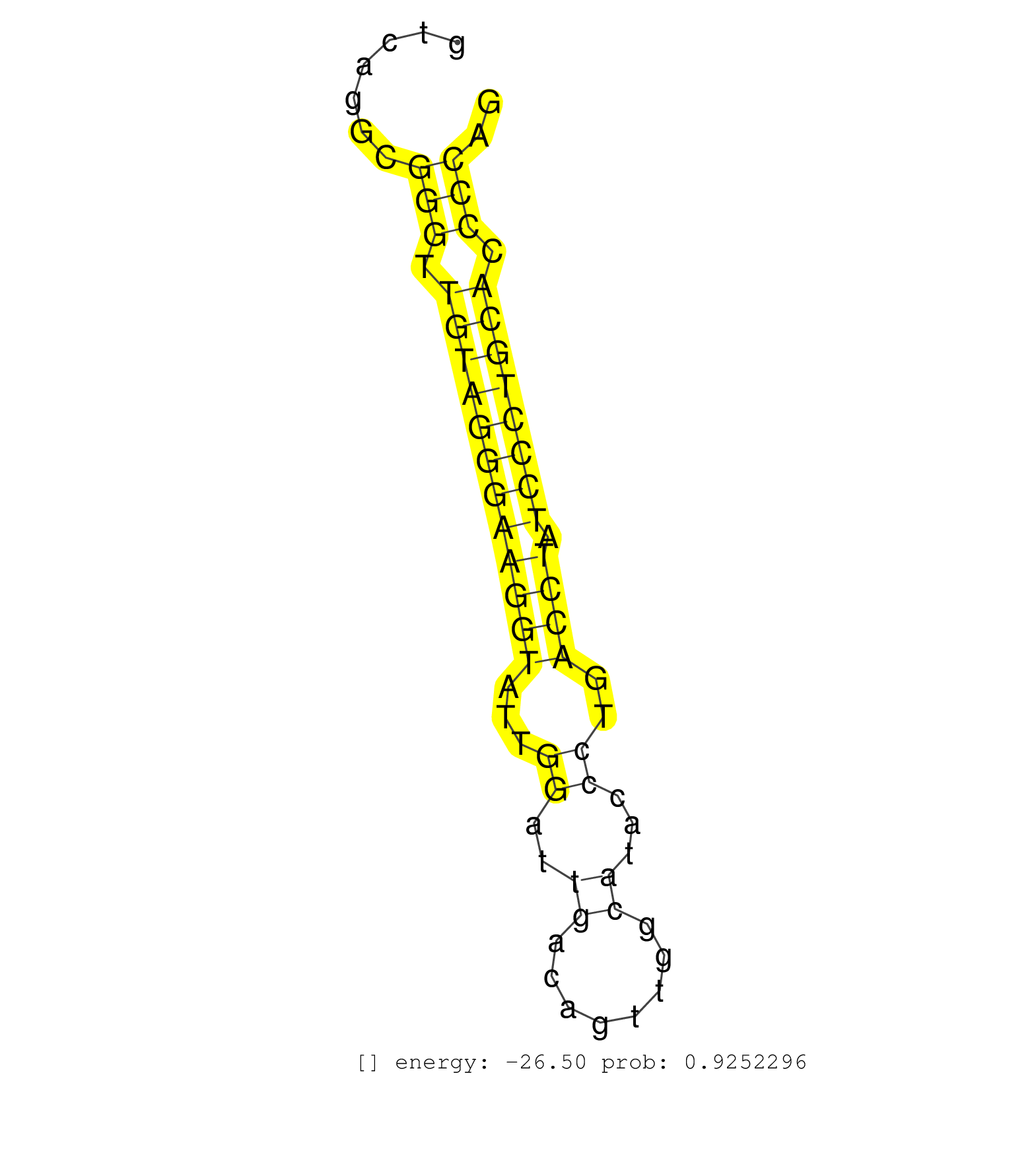
| (1) AGO1.ip | (5) AGO2.ip | (1) AGO3.ip | (24) BRAIN | (1) CELL-LINE | (1) DCR.mut | (1) DGCR8.mut | (6) EMBRYO | (3) ESC | (1) LIVER | (1) LUNG | (8) OTHER | (2) SKIN | (1) TDRD1.ip | (3) TESTES | (8) TOTAL-RNA |
| TAATGATGTTGAGGTGTAAAGTTCTTGGGGGGTCACAGTTTCAGTGGGAAGCTGTGGAAGGCACCTGGGTTGGAGGCTGAGATAGGCAGGATGGGCTCTTGGAGACTCTTTCAGGTATAAGGTGGTTTAGGTGTCAGGCGGGTTGTAGGGAAGGTATTGGATTGACAGTTGGCATACCCTGACCTATCCCTGCACCCCAGTCCTGGGGCTTCCCAACAGCGTGGAGGAGATGTGTCCTGACATCCCAGTG ...........................................................................................................................................(((.((((((((((((...((..((........))...))..)))).)))))))).))).................................................... ....................................................................................................................................133................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR346422(SRX098264) Global profiling of miRNA and the hairpin pre. (Muscle) | SRR553583(SRX182789) source: Cerebellum. (Cerebellum) | mjTestesWT1() Testes Data. (testes) | SRR553582(SRX182788) source: Brain. (Brain) | SRR345203(SRX097264) source: size fractionated RNA from mouse hipp. (brain) | SRR042474(GSM539866) mouse embryonic stem cells [09-002]. (ESC) | SRR346417(SRX098258) Global profiling of miRNA and the hairpin pre. (Brain) | GSM510456(GSM510456) newborn_rep12. (total RNA) | SRR042483(GSM539875) mouse muscle tissue [09-002]. (muscle) | SRR346423(SRX098265) Global profiling of miRNA and the hairpin pre. (Brain) | GSM510452(GSM510452) newborn_rep8. (total RNA) | GSM510454(GSM510454) newborn_rep10. (total RNA) | SRR306529(GSM750572) 19-24nt. (ago2 brain) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) | SRR038744(GSM527279) small RNA-Seq. (brain) | SRR306531(GSM750574) 19-24nt. (ago2 brain) | SRR546155(SRX180174) Global profiling of miRNA and the hairpin pre. (ago2 Neuroblastoma) | SRR306530(GSM750573) 19-24nt. (ago2 brain) | GSM510455(GSM510455) newborn_rep11. (total RNA) | SRR206940(GSM723281) other. (brain) | SRR206939(GSM723280) other. (brain) | SRR116846(GSM678422) Small RNA deep sequencing from mouse epididym. (epididymis) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | SRR023847(GSM307156) mESv6.5smallrna_rep1. (cell line) | SRR037912(GSM510449) newborn_rep5. (total RNA) | SRR345199(SRX097260) source: size fractionated RNA from mouse hipp. (brain) | SRR345202(SRX097263) source: size fractionated RNA from mouse hipp. (brain) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR345201(SRX097262) source: size fractionated RNA from mouse hipp. (brain) | SRR037920(GSM510458) e12p5_rep2. (embryo) | SRR306542(GSM750585) 19-24nt. (ago2 brain) | GSM510450(GSM510450) newborn_rep6. (total RNA) | SRR073954(GSM629280) total RNA. (blood) | Ago3IP812(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR037914(GSM510451) newborn_rep7. (total RNA) | SRR345205(SRX097266) source: size fractionated RNA from mouse hipp. (brain) | SRR042456(GSM539848) mouse hematopoetic progenitor cells [09-002]. (bone marrow) | GSM510453(GSM510453) newborn_rep9. (total RNA) | SRR042477(GSM539869) mouse brain tissue [09-002]. (brain) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR037921(GSM510459) e12p5_rep3. (embryo) | GSM510444(GSM510444) brain_rep5. (brain) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR345207(SRX097268) source: size fractionated RNA from mouse hipp. (brain) | SRR039610(GSM527274) small RNA-Seq. (brain) | Ago1IP517(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | SRR346419(SRX098260) Global profiling of miRNA and the hairpin pre. (Lung) | SRR206942(GSM723283) other. (brain) | SRR038743(GSM527278) small RNA-Seq. (dicer brain) | GSM314557(GSM314557) ESC dgcr8 (Illumina). (ESC) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR039153(GSM471930) HL1_siSrf1_smallRNAseq. (muscle) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR345196(SRX097257) source: size fractionated RNA from mouse hipp. (brain) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................................................TGACCTATCCCTGCACCCCAGT................................................. | 22 | 1 | 202.00 | 202.00 | 173.00 | 3.00 | - | - | - | 1.00 | 1.00 | 3.00 | - | - | 2.00 | 1.00 | 2.00 | - | 2.00 | 2.00 | 1.00 | - | 1.00 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - |
| .........................................................................................................................................GCGGGTTGTAGGGAAGGTATTGG.......................................................................................... | 23 | 1 | 41.00 | 41.00 | 27.00 | 7.00 | - | 2.00 | - | - | 1.00 | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TGACCTATCCCTGCACCCCAGA................................................. | 22 | 1 | 36.00 | 32.00 | 29.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...................................................................................................................................................................................TGACCTATCCCTGCACCCCAG.................................................. | 21 | 1 | 32.00 | 32.00 | 32.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................GCGGGTTGTAGGGAAGGTATTG........................................................................................... | 22 | 1 | 21.00 | 21.00 | 13.00 | 4.00 | - | 1.00 | 1.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................CGGGTTGTAGGGAAGGTATTGG.......................................................................................... | 22 | 1 | 20.00 | 20.00 | 10.00 | - | - | - | - | 3.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................CTGACCTATCCCTGCACCCCAGT................................................. | 23 | 1 | 20.00 | 20.00 | 13.00 | 1.00 | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................CGGGTTGTAGGGAAGGTATTGGA......................................................................................... | 23 | 1 | 16.00 | 16.00 | 11.00 | - | 4.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................GCGGGTTGTAGGGAAGGTATT............................................................................................ | 21 | 1 | 10.00 | 10.00 | 3.00 | - | - | 3.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................GCGGGTTGTAGGGAAGGTATTGGAT........................................................................................ | 25 | 1 | 10.00 | 10.00 | 4.00 | - | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................CTGACCTATCCCTGCACCCCAG.................................................. | 22 | 1 | 9.00 | 9.00 | 7.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TGACCTATCCCTGCACCCCAGTT................................................ | 23 | 1 | 8.00 | 202.00 | 8.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................GGCGGGTTGTAGGGAAGGTATTGG.......................................................................................... | 24 | 1 | 7.00 | 7.00 | 3.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TGACCTATCCCTGCACCCCAGTA................................................ | 23 | 1 | 7.00 | 202.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TGACCTATCCCTGCACCCCAGAA................................................ | 23 | 1 | 6.00 | 32.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TGACCTATCCCTGCACCCCATT................................................. | 22 | 1 | 5.00 | 2.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................CGGGTTGTAGGGAAGGTATTGGT......................................................................................... | 23 | 1 | 5.00 | 20.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .........................................................................................................................................GCGGGTTGTAGGGAAGGTATTGGA......................................................................................... | 24 | 1 | 5.00 | 5.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TGACCTATCCCTGCACCCCAGAT................................................ | 23 | 1 | 4.00 | 32.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................CGGGTTGTAGGGAAGGTATTGGAT........................................................................................ | 24 | 1 | 4.00 | 4.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TGACCTATCCCTGCACCCC.................................................... | 19 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TGACCTATCCCTGCACCCCAGC................................................. | 22 | 1 | 3.00 | 32.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................CCTGACCTATCCCTGCACCCCAT.................................................. | 23 | 3.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................CGGGTTGTAGGGAAGGTATT............................................................................................ | 20 | 1 | 3.00 | 3.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................GGCGGGTTGTAGGGAAGGTATTG........................................................................................... | 23 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................GCGGGTTGTAGGGAAGGTATTGGT......................................................................................... | 24 | 1 | 3.00 | 41.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TGACCTATCCCTGCACCCCAGTTT............................................... | 24 | 1 | 2.00 | 202.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................CTGACCTATCCCTGCACCCCAGTT................................................ | 24 | 1 | 2.00 | 20.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................GCGGGTTGTAGGGAAGGTAT............................................................................................. | 20 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TGACCTATCCCTGCACCCCA................................................... | 20 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................CCTGACCTATCCCTGCACCCCAGT................................................. | 24 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................CTGACCTATCCCTGCACCCCAGA................................................. | 23 | 1 | 2.00 | 9.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................CGGGTTGTAGGGAAGGTATTGGAAA....................................................................................... | 25 | 1 | 1.00 | 16.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................GGGTTGTAGGGAAGGTATTGG.......................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................CTGACCTATCCCTGCACCCCAGTTT............................................... | 25 | 1 | 1.00 | 20.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................GCGGGTTGTAGGGAAGGTATTGT.......................................................................................... | 23 | 1 | 1.00 | 21.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................CTGACCTATCCCTGCACCCC.................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................CCTGACCTATCCCTGCACCCC.................................................... | 21 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................GCGGGTTGTAGGGAAGGTATTGGTAT....................................................................................... | 26 | 1 | 1.00 | 41.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................CTGACCTATCCCTGCACCCCAT.................................................. | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................GAAGGCACCTGGGTTCACC............................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................................ATTGGATTGACAGTTGGC............................................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................TGGGTTGGAGGCTGAGGA....................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | |
| .................................................................................................................................................................................CCTGACCTATCCCTGCACCCCAGA................................................. | 24 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................TGACCTATCCCTGCACCCCAGAGA............................................... | 24 | 1 | 1.00 | 32.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................GCGGGTTGTAGGGAAGGTATTGA.......................................................................................... | 23 | 1 | 1.00 | 21.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TGACCTATCCCTGCACCCCAGTAA............................................... | 24 | 1 | 1.00 | 202.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................GGGAAGCTGTGGAAGCTCT.......................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................CGGGTTGTAGGGAAGGTATTGA.......................................................................................... | 22 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................CGGGTTGTAGGGAAGGTATTGTA......................................................................................... | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................CGGGTTGTAGGGAAGGAATT............................................................................................ | 20 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................................GCGGGTTGTAGGGAAGGTATTGGAAT....................................................................................... | 26 | 1 | 1.00 | 5.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................CTGACCTATCCCTGCACCCCA................................................... | 21 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........TGAGGTGTAAAGTTCTGCGG............................................................................................................................................................................................................................. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................TGACCTATCCCTGCACCCCAGTAT............................................... | 24 | 1 | 1.00 | 202.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................GCGGGTTGTAGGGAAGGTATTGGATT....................................................................................... | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................GCGGGTTGTAGGGAAGGTA.............................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................TTTCAGGTATAAGGTGGGA........................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................CTGACCTATCCCTGCACCCCAGTAT............................................... | 25 | 1 | 1.00 | 20.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................GCGGGTTGTAGGGAAGGTATTTG.......................................................................................... | 23 | 1 | 1.00 | 10.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................GCGGGTTGTAGGGAAGGGATT............................................................................................ | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................TGGATTGACAGTTGGTTA........................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | |
| ...................................................................................................................................................................................TGACCTATCCCTGCACCCCACC................................................. | 22 | 1 | 1.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................GTGTCAGGCGGGTTG......................................................................................................... | 15 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................CGTGGAGGAGATGTGT............... | 16 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 |
| TAATGATGTTGAGGTGTAAAGTTCTTGGGGGGTCACAGTTTCAGTGGGAAGCTGTGGAAGGCACCTGGGTTGGAGGCTGAGATAGGCAGGATGGGCTCTTGGAGACTCTTTCAGGTATAAGGTGGTTTAGGTGTCAGGCGGGTTGTAGGGAAGGTATTGGATTGACAGTTGGCATACCCTGACCTATCCCTGCACCCCAGTCCTGGGGCTTCCCAACAGCGTGGAGGAGATGTGTCCTGACATCCCAGTG ...........................................................................................................................................(((.((((((((((((...((..((........))...))..)))).)))))))).))).................................................... ....................................................................................................................................133................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR346422(SRX098264) Global profiling of miRNA and the hairpin pre. (Muscle) | SRR553583(SRX182789) source: Cerebellum. (Cerebellum) | mjTestesWT1() Testes Data. (testes) | SRR553582(SRX182788) source: Brain. (Brain) | SRR345203(SRX097264) source: size fractionated RNA from mouse hipp. (brain) | SRR042474(GSM539866) mouse embryonic stem cells [09-002]. (ESC) | SRR346417(SRX098258) Global profiling of miRNA and the hairpin pre. (Brain) | GSM510456(GSM510456) newborn_rep12. (total RNA) | SRR042483(GSM539875) mouse muscle tissue [09-002]. (muscle) | SRR346423(SRX098265) Global profiling of miRNA and the hairpin pre. (Brain) | GSM510452(GSM510452) newborn_rep8. (total RNA) | GSM510454(GSM510454) newborn_rep10. (total RNA) | SRR306529(GSM750572) 19-24nt. (ago2 brain) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) | SRR038744(GSM527279) small RNA-Seq. (brain) | SRR306531(GSM750574) 19-24nt. (ago2 brain) | SRR546155(SRX180174) Global profiling of miRNA and the hairpin pre. (ago2 Neuroblastoma) | SRR306530(GSM750573) 19-24nt. (ago2 brain) | GSM510455(GSM510455) newborn_rep11. (total RNA) | SRR206940(GSM723281) other. (brain) | SRR206939(GSM723280) other. (brain) | SRR116846(GSM678422) Small RNA deep sequencing from mouse epididym. (epididymis) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | SRR023847(GSM307156) mESv6.5smallrna_rep1. (cell line) | SRR037912(GSM510449) newborn_rep5. (total RNA) | SRR345199(SRX097260) source: size fractionated RNA from mouse hipp. (brain) | SRR345202(SRX097263) source: size fractionated RNA from mouse hipp. (brain) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR345201(SRX097262) source: size fractionated RNA from mouse hipp. (brain) | SRR037920(GSM510458) e12p5_rep2. (embryo) | SRR306542(GSM750585) 19-24nt. (ago2 brain) | GSM510450(GSM510450) newborn_rep6. (total RNA) | SRR073954(GSM629280) total RNA. (blood) | Ago3IP812(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR037914(GSM510451) newborn_rep7. (total RNA) | SRR345205(SRX097266) source: size fractionated RNA from mouse hipp. (brain) | SRR042456(GSM539848) mouse hematopoetic progenitor cells [09-002]. (bone marrow) | GSM510453(GSM510453) newborn_rep9. (total RNA) | SRR042477(GSM539869) mouse brain tissue [09-002]. (brain) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR037921(GSM510459) e12p5_rep3. (embryo) | GSM510444(GSM510444) brain_rep5. (brain) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR345207(SRX097268) source: size fractionated RNA from mouse hipp. (brain) | SRR039610(GSM527274) small RNA-Seq. (brain) | Ago1IP517(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | SRR346419(SRX098260) Global profiling of miRNA and the hairpin pre. (Lung) | SRR206942(GSM723283) other. (brain) | SRR038743(GSM527278) small RNA-Seq. (dicer brain) | GSM314557(GSM314557) ESC dgcr8 (Illumina). (ESC) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR039153(GSM471930) HL1_siSrf1_smallRNAseq. (muscle) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR345196(SRX097257) source: size fractionated RNA from mouse hipp. (brain) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................................CCTGACCTATCCCTGCCGG...................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................GGGGTCACAGTTTCAGTT............................................................................................................................................................................................................ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................................................................CCTGCACCCCAGTCCTGT............................................ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................................................................CTATCCCTGCACCCCAAG................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................................................TCCCAACAGCGTGG.......................... | 14 | 7 | 0.14 | 0.14 | - | - | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |