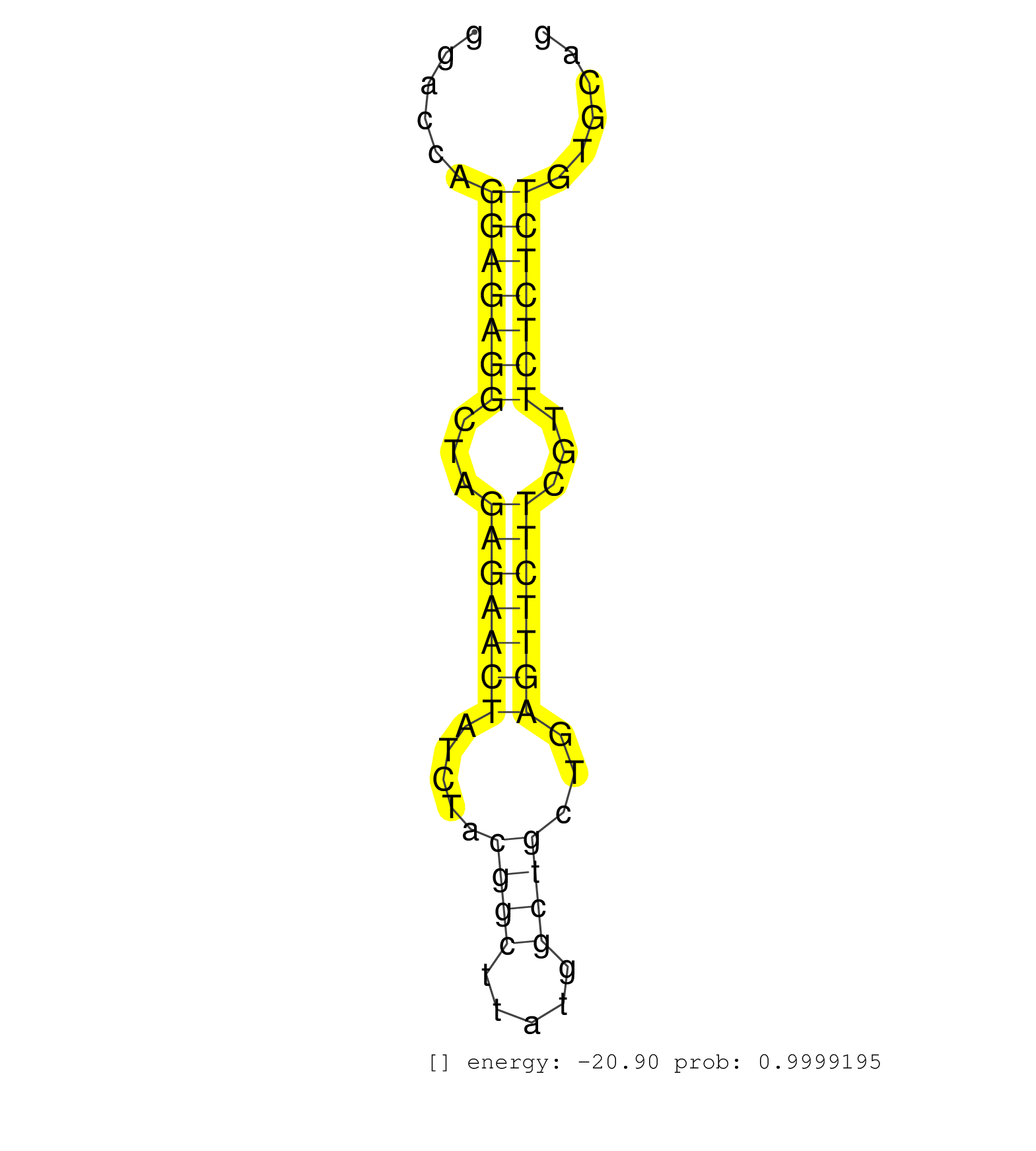
| (9) AGO2.ip | (30) BRAIN | (2) DCR.mut | (2) DGCR8.mut | (2) ESC | (1) KIDNEY | (1) OTHER | (1) PIWI.mut | (1) TESTES | (2) TOTAL-RNA |
| GGAAAGTTGAGAACCGTTGTGTTAGATAAAGTCTTTTTATTTTCAGCATATGTTCTAGAAACTATTCTTTCCATCGTGGTCTGTGCTGGAATTGTTTTGCCCTTGCTTAACAGCTTCTAAGACAATTCAGAGAGGACCAGGAGAGGCTAGAGAACTATCTACGGCTTATGGCTGCTGAGTTCTTCGTTCTCTCTGTGCAGGTGGGAAAATGGAAAGACAAGTCCCTGCAGATGAAATACTATGTGTGGCC ...........................................................................................................................................(((((((...(((((((.....((((.....))))...)))))))...)))))))........................................................ .....................................................................................................................................134...............................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR206940(GSM723281) other. (brain) | SRR346423(SRX098265) Global profiling of miRNA and the hairpin pre. (Brain) | SRR553582(SRX182788) source: Brain. (Brain) | SRR306533(GSM750576) 19-24nt. (ago2 brain) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) | SRR206939(GSM723280) other. (brain) | SRR345201(SRX097262) source: size fractionated RNA from mouse hipp. (brain) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR038741(GSM527276) small RNA-Seq. (brain) | SRR345207(SRX097268) source: size fractionated RNA from mouse hipp. (brain) | SRR345205(SRX097266) source: size fractionated RNA from mouse hipp. (brain) | SRR345203(SRX097264) source: size fractionated RNA from mouse hipp. (brain) | SRR306542(GSM750585) 19-24nt. (ago2 brain) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR306534(GSM750577) 19-24nt. (ago2 brain) | SRR306540(GSM750583) 19-24nt. (ago2 brain) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR042477(GSM539869) mouse brain tissue [09-002]. (brain) | SRR038744(GSM527279) small RNA-Seq. (brain) | SRR306536(GSM750579) 19-24nt. (ago2 brain) | SRR038745(GSM527280) small RNA-Seq. (dgcr8 brain) | SRR279906(GSM689056) cell type: mouse embryonic stem cellcell line. (ESC) | SRR038743(GSM527278) small RNA-Seq. (dicer brain) | SRR206942(GSM723283) other. (brain) | SRR306537(GSM750580) 19-24nt. (ago2 brain) | SRR306543(GSM750586) 19-24nt. (ago2 brain) | SRR345204(SRX097265) source: size fractionated RNA from mouse hipp. (brain) | SRR206941(GSM723282) other. (brain) | SRR306535(GSM750578) 19-24nt. (ago2 brain) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR553583(SRX182789) source: Cerebellum. (Cerebellum) | SRR037912(GSM510449) newborn_rep5. (total RNA) | SRR038740(GSM527275) small RNA-Seq. (dicer brain) | SRR306532(GSM750575) 19-24nt. (ago2 brain) | GSM510456(GSM510456) newborn_rep12. (total RNA) | SRR346416(SRX098257) Global profiling of miRNA and the hairpin pre. (Kidney) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................................TGAGTTCTTCGTTCTCTCTGTGC.................................................... | 23 | 1 | 60.00 | 60.00 | 15.00 | 6.00 | 7.00 | 2.00 | - | 7.00 | - | 5.00 | 6.00 | - | - | - | 3.00 | - | 1.00 | - | - | - | 2.00 | 1.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | - |
| ..........................................................................................................................................AGGAGAGGCTAGAGAACTATCT.......................................................................................... | 22 | 1 | 17.00 | 17.00 | - | - | - | - | 3.00 | - | - | - | - | 4.00 | 2.00 | 2.00 | - | - | - | - | 1.00 | 3.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................TGAGTTCTTCGTTCTCTCTGTG..................................................... | 22 | 1 | 10.00 | 10.00 | - | - | - | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | 1.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................AGGAGAGGCTAGAGAACTAT............................................................................................ | 20 | 1 | 9.00 | 9.00 | - | - | - | - | 4.00 | - | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................TGAGTTCTTCGTTCTCTCTGTGT.................................................... | 23 | 1 | 8.00 | 10.00 | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | 1.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................TGAGTTCTTCGTTCTCTCTGTGA.................................................... | 23 | 1 | 6.00 | 10.00 | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................TGAGTTCTTCGTTCTCTCTGT...................................................... | 21 | 1 | 3.00 | 3.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................AGGAGAGGCTAGAGAACTATCC.......................................................................................... | 22 | 1 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................AGGAGAGGCTAGAGAACTCTCT.......................................................................................... | 22 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................................................TGAGTTCTTCGTTCTCTCTGTT..................................................... | 22 | 1 | 2.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...............................................................................................................................................................................TGAGTTCTTCGTTCTCTCTGTGCA................................................... | 24 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TTCTTCGTTCTCTCTGTGCAGT................................................. | 22 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................................................TGAGTTCTTCGTTCTCTCTGCGC.................................................... | 23 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................AGGAGAGGCTAGAGAACTCT............................................................................................ | 20 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................................................TGAGTTCTTCGTTCTCTCTGTTG.................................................... | 23 | 1 | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................TTAACAGCTTCTAAGACCC............................................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | |
| ...........AACCGTTGTGTTAGACTGA............................................................................................................................................................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................................................TGAGTTCTTCGTTCTCTCTGAAA.................................................... | 23 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................AGGAGAGGCTAGAGATGGC............................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................AGGAGAGGCTAGAGAACTATC........................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................AGGAGAGGCTAGAGAACTAG............................................................................................ | 20 | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................................................GAAAGACAAGTCCCTGCAGATGAAAT............. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................TGAGTTCTTCGTTCTCTCTTTGC.................................................... | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................................................TGAGTTCTTCGTTCTCTCTGTTT.................................................... | 23 | 1 | 1.00 | 3.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................CTGAGTTCTTCGTTCTCTCTGTGC.................................................... | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................AAAATGGAAAGACAACGCC.......................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | |
| .......................................................................................................................................ACCAGGAGAGGCTAGAGAAC............................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................GAGGACCAGGAGAGGC....................................................................................................... | 16 | 5 | 0.40 | 0.40 | - | - | - | - | - | - | - | - | - | - | 0.40 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GGAAAGTTGAGAACCGTTGTGTTAGATAAAGTCTTTTTATTTTCAGCATATGTTCTAGAAACTATTCTTTCCATCGTGGTCTGTGCTGGAATTGTTTTGCCCTTGCTTAACAGCTTCTAAGACAATTCAGAGAGGACCAGGAGAGGCTAGAGAACTATCTACGGCTTATGGCTGCTGAGTTCTTCGTTCTCTCTGTGCAGGTGGGAAAATGGAAAGACAAGTCCCTGCAGATGAAATACTATGTGTGGCC ...........................................................................................................................................(((((((...(((((((.....((((.....))))...)))))))...)))))))........................................................ .....................................................................................................................................134...............................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR206940(GSM723281) other. (brain) | SRR346423(SRX098265) Global profiling of miRNA and the hairpin pre. (Brain) | SRR553582(SRX182788) source: Brain. (Brain) | SRR306533(GSM750576) 19-24nt. (ago2 brain) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) | SRR206939(GSM723280) other. (brain) | SRR345201(SRX097262) source: size fractionated RNA from mouse hipp. (brain) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR038741(GSM527276) small RNA-Seq. (brain) | SRR345207(SRX097268) source: size fractionated RNA from mouse hipp. (brain) | SRR345205(SRX097266) source: size fractionated RNA from mouse hipp. (brain) | SRR345203(SRX097264) source: size fractionated RNA from mouse hipp. (brain) | SRR306542(GSM750585) 19-24nt. (ago2 brain) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR306534(GSM750577) 19-24nt. (ago2 brain) | SRR306540(GSM750583) 19-24nt. (ago2 brain) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR042477(GSM539869) mouse brain tissue [09-002]. (brain) | SRR038744(GSM527279) small RNA-Seq. (brain) | SRR306536(GSM750579) 19-24nt. (ago2 brain) | SRR038745(GSM527280) small RNA-Seq. (dgcr8 brain) | SRR279906(GSM689056) cell type: mouse embryonic stem cellcell line. (ESC) | SRR038743(GSM527278) small RNA-Seq. (dicer brain) | SRR206942(GSM723283) other. (brain) | SRR306537(GSM750580) 19-24nt. (ago2 brain) | SRR306543(GSM750586) 19-24nt. (ago2 brain) | SRR345204(SRX097265) source: size fractionated RNA from mouse hipp. (brain) | SRR206941(GSM723282) other. (brain) | SRR306535(GSM750578) 19-24nt. (ago2 brain) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR553583(SRX182789) source: Cerebellum. (Cerebellum) | SRR037912(GSM510449) newborn_rep5. (total RNA) | SRR038740(GSM527275) small RNA-Seq. (dicer brain) | SRR306532(GSM750575) 19-24nt. (ago2 brain) | GSM510456(GSM510456) newborn_rep12. (total RNA) | SRR346416(SRX098257) Global profiling of miRNA and the hairpin pre. (Kidney) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................GTCTTTTTATTTTCAGA........................................................................................................................................................................................................... | 17 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................TTCTTCGTTCTCTCTGACT.................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................TTGTTTTGCCCTTGCGAC............................................................................................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | |
| ............................................AGCATATGTTCTAGAAC............................................................................................................................................................................................. | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................................AGTTCTTCGTTCTCTC......................................................... | 16 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - |
| ...........................................................................................................................................................TATCTACGGCTTA.................................................................................. | 13 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 |