| (1) AGO.mut | (3) AGO2.ip | (5) BRAIN | (4) CELL-LINE | (9) EMBRYO | (1) ESC | (3) FIBROBLAST | (2) LIVER | (4) OTHER | (2) OTHER.mut | (1) SPLEEN | (2) TESTES | (1) THYMUS | (11) TOTAL-RNA | (1) UTERUS |
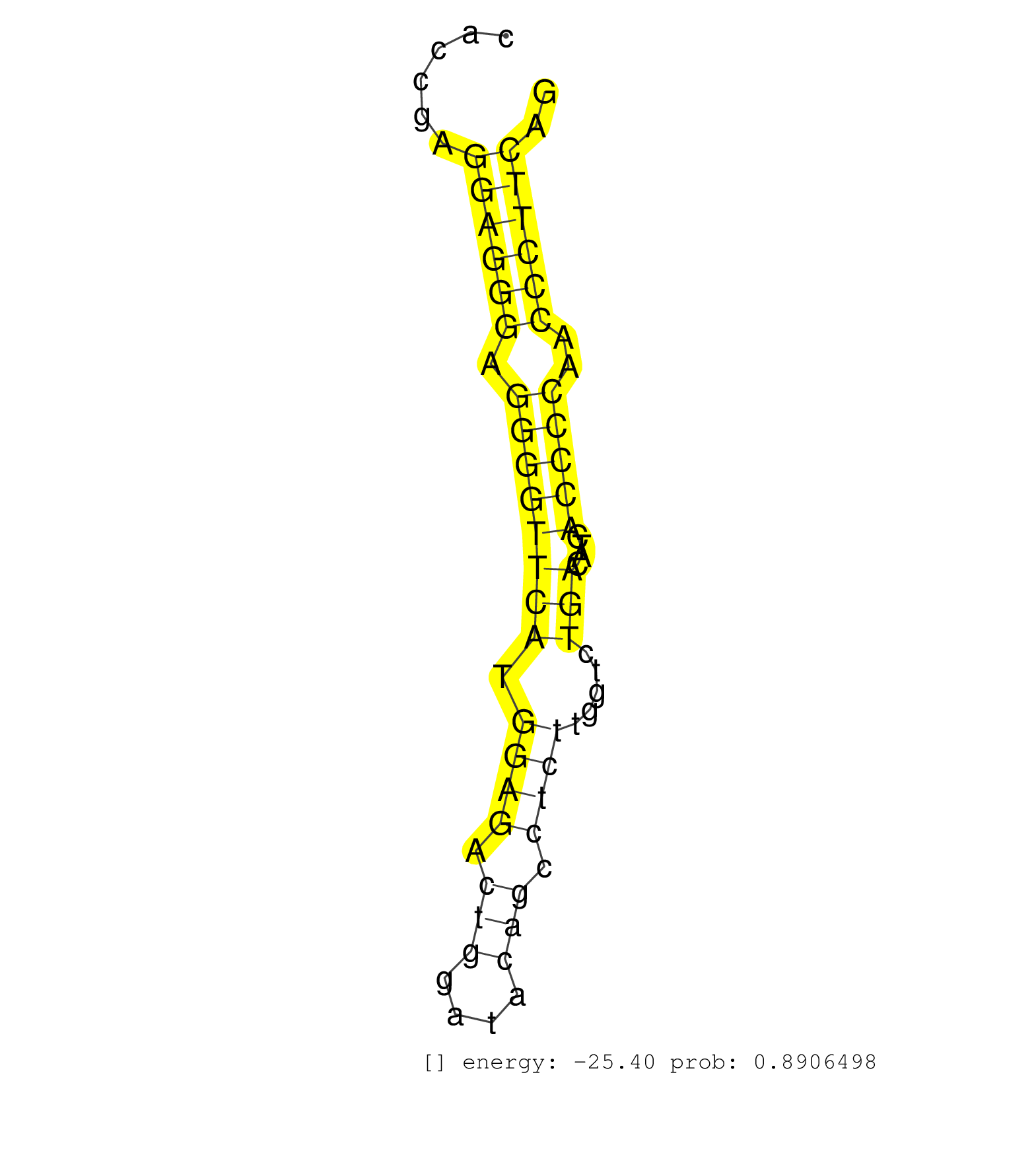
| AGGCCGGGAACGCCTGGGCTCCTTCGGCAGCATCACCCGGCAGCAGGAAGGTGAGCAGAGAATACTGGGAACTGGGGTCCTCACAGGGAGGCTGGGGGGCCACCTGTTCTGTGAGTCAAAATGGCCTCATCCCTGACCACCTTGGCCCAGATGGCCAGTCAGGCCTGAGCAGAGAAGGGGGTTCACCGAGGAGGGAGGGGTTCATGGAGACTGGATACAGCCTCTTGGTCTGACATCACCCCAACCCTTCAGGTGAGGCCAGCTCTCAGGACATGACAGCCCAGGTGACAAGTCCGTCTGGC .............................................................................................................................................................................................((((((.((((((((.((((.(((....))).)))).....)))....)))))..)))))).................................................... .......................................................................................................................................................................................184.................................................................252................................................ | Size | Perfect hit | Total Norm | Perfect Norm | GSM510456(GSM510456) newborn_rep12. (total RNA) | SRR346422(SRX098264) Global profiling of miRNA and the hairpin pre. (Muscle) | SRR546155(SRX180174) Global profiling of miRNA and the hairpin pre. (ago2 Neuroblastoma) | SRR685339(GSM1079783) Small RNAs (15-50 nts in length) from immorta. (cell line) | SRR039183(GSM485323) shLuc.1309 (-dox). (fibroblast) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR037914(GSM510451) newborn_rep7. (total RNA) | GSM510454(GSM510454) newborn_rep10. (total RNA) | GSM510450(GSM510450) newborn_rep6. (total RNA) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | GSM510452(GSM510452) newborn_rep8. (total RNA) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR037912(GSM510449) newborn_rep5. (total RNA) | GSM510453(GSM510453) newborn_rep9. (total RNA) | SRR039185(GSM485325) shLuc.1309;ROSA-rtTA (-dox). (fibroblast) | SRR023851(GSM307160) ZHBT-c412hsmallrna_rep1. (cell line) | SRR345207(SRX097268) source: size fractionated RNA from mouse hipp. (brain) | SRR390300(GSM849858) cell line: NIH-3T3cell type: fibroblastinfect. (ago2 fibroblast) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (ago2 liver) | SRR346414(SRX098255) Global profiling of miRNA and the hairpin pre. (Spleen) | SRR553602(SRX182808) source: Sertoli cells. (Sertoli cells) | SRR037911(GSM510448) newborn_rep4. (total RNA) | SRR037909(GSM510446) newborn_rep2. (total RNA) | SRR346420(SRX098261) Global profiling of miRNA and the hairpin pre. (Uterus) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | SRR059765(GSM562827) DN3_control. (thymus) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR345196(SRX097257) source: size fractionated RNA from mouse hipp. (brain) | SRR037925(GSM510463) e9p5_rep3. (embryo) | SRR037908(GSM510445) newborn_rep1. (total RNA) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | GSM510455(GSM510455) newborn_rep11. (total RNA) | SRR037924(GSM510462) e9p5_rep2. (embryo) | SRR037937(GSM510475) 293cand2. (cell line) | SRR346425(SRX098267) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR037927(GSM510465) e7p5_rep1. (embryo) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................................................................................................TGACATCACCCCAACCCTTCAGT................................................. | 23 | 1 | 38.00 | 12.00 | 7.00 | 3.00 | 4.00 | - | - | - | 3.00 | - | 2.00 | 1.00 | - | 2.00 | 2.00 | 2.00 | 2.00 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................................TGACATCACCCCAACCCTTCAG.................................................. | 22 | 1 | 12.00 | 12.00 | - | 7.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - |
| ....................................................................................................................................................................................................................................TCTGACATCACCCCAACCCTTCAGT................................................. | 25 | 9.00 | 0.00 | 6.00 | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | |
| ............................................................................................................................................................................................AGGAGGGAGGGGTTCATGGAGA............................................................................................ | 22 | 1 | 5.00 | 5.00 | - | - | - | 4.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................................TGACATCACCCCAACCCTTCAGA................................................. | 23 | 1 | 5.00 | 12.00 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...........................................................GAATACTGGGAACTGGGGT................................................................................................................................................................................................................................ | 19 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................................TCTGACATCACCCCATAA........................................................ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | |
| ......................TTCGGCAGCATCACCCGGCA.................................................................................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................GCAGGAAGGTGAGCAGGGA................................................................................................................................................................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................................................................................................................GACAGCCCAGGTGACAAGTCCGTCT... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................................TGACATCACCCCAACCCTTCAGTA................................................ | 24 | 1 | 1.00 | 12.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................GCAGCATCACCCGGCT.................................................................................................................................................................................................................................................................... | 16 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | |
| ..........................................GCAGGAAGGTGAGCAGAGCG................................................................................................................................................................................................................................................ | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................................................................................................TGACATCACCCCAACCCTTCATA................................................. | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................................................................................................................................CAGGACATGACAGCCCAGG................. | 19 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | 0.50 |
| ...............................ATCACCCGGCAGCAGGAAGGT.......................................................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................TGAGGCCAGCTCTCAGGACA............................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................TCACAGGGAGGCTGGTTGG........................................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................................................................CACCGAGGAGGGAGGCAG..................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AGGCCGGGAACGCCTGGGCTCCTTCGGCAGCATCACCCGGCAGCAGGAAGGTGAGCAGAGAATACTGGGAACTGGGGTCCTCACAGGGAGGCTGGGGGGCCACCTGTTCTGTGAGTCAAAATGGCCTCATCCCTGACCACCTTGGCCCAGATGGCCAGTCAGGCCTGAGCAGAGAAGGGGGTTCACCGAGGAGGGAGGGGTTCATGGAGACTGGATACAGCCTCTTGGTCTGACATCACCCCAACCCTTCAGGTGAGGCCAGCTCTCAGGACATGACAGCCCAGGTGACAAGTCCGTCTGGC .............................................................................................................................................................................................((((((.((((((((.((((.(((....))).)))).....)))....)))))..)))))).................................................... .......................................................................................................................................................................................184.................................................................252................................................ | Size | Perfect hit | Total Norm | Perfect Norm | GSM510456(GSM510456) newborn_rep12. (total RNA) | SRR346422(SRX098264) Global profiling of miRNA and the hairpin pre. (Muscle) | SRR546155(SRX180174) Global profiling of miRNA and the hairpin pre. (ago2 Neuroblastoma) | SRR685339(GSM1079783) Small RNAs (15-50 nts in length) from immorta. (cell line) | SRR039183(GSM485323) shLuc.1309 (-dox). (fibroblast) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR037914(GSM510451) newborn_rep7. (total RNA) | GSM510454(GSM510454) newborn_rep10. (total RNA) | GSM510450(GSM510450) newborn_rep6. (total RNA) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | GSM510452(GSM510452) newborn_rep8. (total RNA) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR037912(GSM510449) newborn_rep5. (total RNA) | GSM510453(GSM510453) newborn_rep9. (total RNA) | SRR039185(GSM485325) shLuc.1309;ROSA-rtTA (-dox). (fibroblast) | SRR023851(GSM307160) ZHBT-c412hsmallrna_rep1. (cell line) | SRR345207(SRX097268) source: size fractionated RNA from mouse hipp. (brain) | SRR390300(GSM849858) cell line: NIH-3T3cell type: fibroblastinfect. (ago2 fibroblast) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (ago2 liver) | SRR346414(SRX098255) Global profiling of miRNA and the hairpin pre. (Spleen) | SRR553602(SRX182808) source: Sertoli cells. (Sertoli cells) | SRR037911(GSM510448) newborn_rep4. (total RNA) | SRR037909(GSM510446) newborn_rep2. (total RNA) | SRR346420(SRX098261) Global profiling of miRNA and the hairpin pre. (Uterus) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | SRR059765(GSM562827) DN3_control. (thymus) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR345196(SRX097257) source: size fractionated RNA from mouse hipp. (brain) | SRR037925(GSM510463) e9p5_rep3. (embryo) | SRR037908(GSM510445) newborn_rep1. (total RNA) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | GSM510455(GSM510455) newborn_rep11. (total RNA) | SRR037924(GSM510462) e9p5_rep2. (embryo) | SRR037937(GSM510475) 293cand2. (cell line) | SRR346425(SRX098267) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR037927(GSM510465) e7p5_rep1. (embryo) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................................................................................................................................CAGCTCTCAGGACATACGA........................ | 19 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................GGCCTCATCCCTGACCTGGA................................................................................................................................................................ | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................................GGGGTTCACCGAGGATTAT.......................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | |
| .............................................................................................................................CTCATCCCTGACCACCAGCA............................................................................................................................................................. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................CTGTTCTGTGAGTCAATA..................................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................................GGGGTTCACCGAGGAT............................................................................................................. | 16 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................................GGGGTTCACCGAGGATTTT.......................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |