| (2) AGO1.ip | (1) AGO2.ip | (1) AGO3.ip | (17) BRAIN | (3) CELL-LINE | (1) DCR.mut | (2) DGCR8.mut | (8) EMBRYO | (2) ESC | (1) KIDNEY | (3) LIVER | (2) LUNG | (8) OTHER | (1) OTHER.mut | (1) OVARY | (4) SKIN | (6) TESTES | (5) TOTAL-RNA |
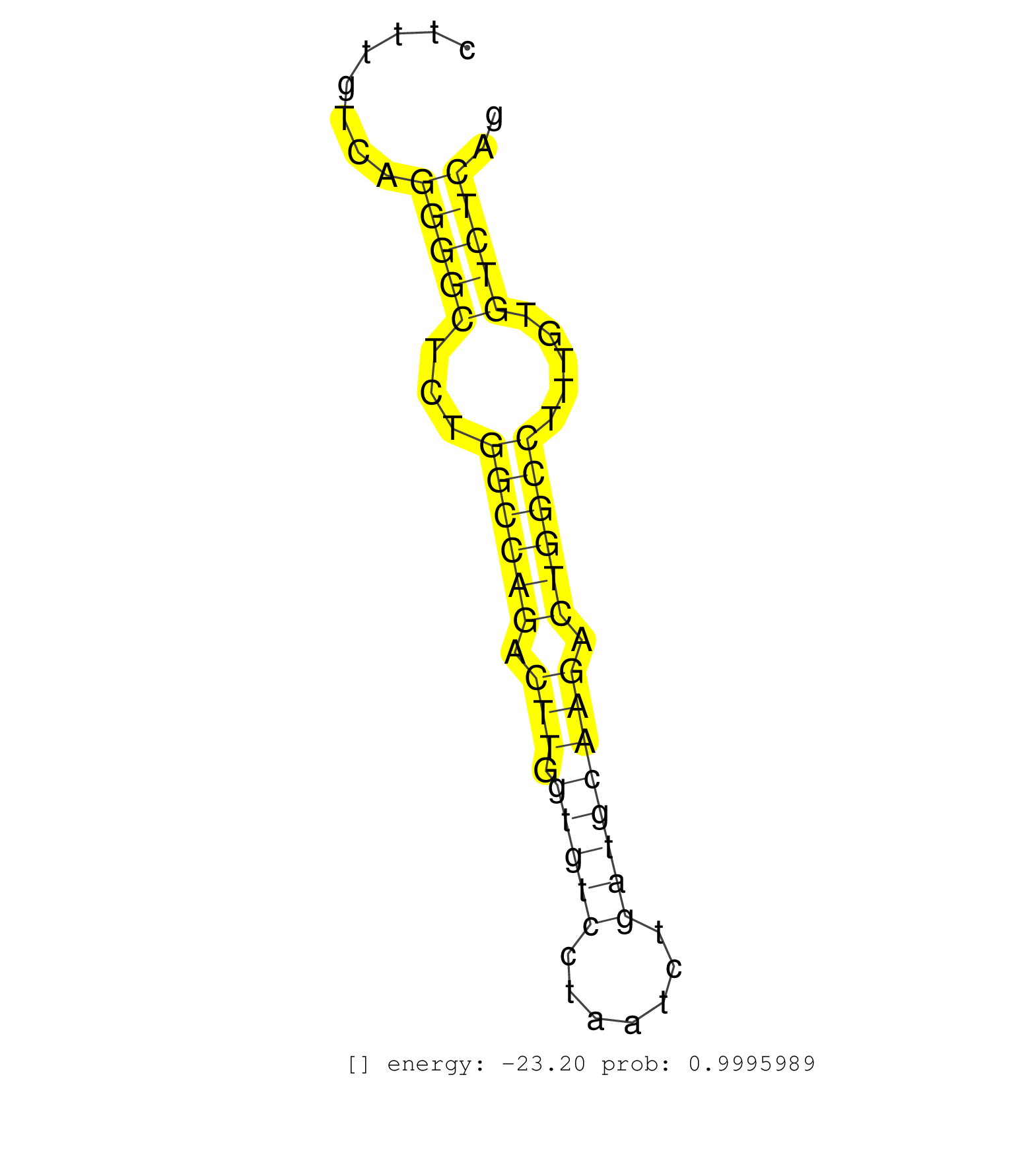
| TGCCCAGCAGGAGCAATGTTTACACAACCTTGAAACAGGAGATACTTAGAGTAAGCATTACAGTGTCACATCCATATTGCACCAAGATATAGGGGTGGAGAAACCAGCATTTGAATGATACAAGTGCACAACACATTCTGCCTACACTGTCCCAGCCCGGGAGGAGAGGAACGGGGTCCTTAGGCTCCCTGTATGCTTCCAAGATTTGCGGATGGGCTGTGAGTGGGTCAGGGTAGCTGGCCTTAGAGACAGCCCTAGTGAGATCCTACATGGATCCTTAAGACAAGGTCTGGGACCCTTACCTCCCATCTGGGGAGAGGGTAGGAGGAGTGTAGGGTAAGGTCAGCTGTCCCCACTGTTGAGAGTGGAGGAGCTTCTTTGTCAGGGGCTCTGGCCAGACTTGGTGTCCTAATCTGATGCAAGACTGGCCTTTGTGTCTCAGGGTGTCGCCAAGGCTACTTTGGGCCCGGCTGCGAGCAGAAGTGCAGGTGT ................................................................................................................................................................................................................................................................................................................................................................................................(((((...((((((.(((.(((((.......)))))))).)))))).....))))).................................................... ........................................................................................................................................................................................................................................................................................................................................................................................377..............................................................442................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | mjLiverWT1() Liver Data. (liver) | SRR345204(SRX097265) source: size fractionated RNA from mouse hipp. (brain) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | mjTestesWT4() Testes Data. (testes) | SRR685339(GSM1079783) Small RNAs (15-50 nts in length) from immorta. (cell line) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR306536(GSM750579) 19-24nt. (ago2 brain) | SRR279904(GSM689054) cell type: mouse embryonic stem cellcell line. (ESC) | SRR346419(SRX098260) Global profiling of miRNA and the hairpin pre. (Lung) | SRR206940(GSM723281) other. (brain) | SRR037937(GSM510475) 293cand2. (cell line) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR345201(SRX097262) source: size fractionated RNA from mouse hipp. (brain) | GSM510456(GSM510456) newborn_rep12. (total RNA) | SRR346423(SRX098265) Global profiling of miRNA and the hairpin pre. (Brain) | SRR037914(GSM510451) newborn_rep7. (total RNA) | RuiDGCR8KO(Rui) DGCR8-dependent microRNA biogenesis is essent. (dgcr8 skin) | SRR345199(SRX097260) source: size fractionated RNA from mouse hipp. (brain) | SRR206941(GSM723282) other. (brain) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) | SRR073954(GSM629280) total RNA. (blood) | SRR065050(SRR065050) Tissue-specific Regulation of Mouse MicroRNA . (liver) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | GSM510455(GSM510455) newborn_rep11. (total RNA) | SRR345206(SRX097267) source: size fractionated RNA from mouse hipp. (brain) | SRR037905(GSM510441) brain_rep2. (brain) | GSM510454(GSM510454) newborn_rep10. (total RNA) | SRR073955(GSM629281) total RNA. (blood) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR346421(SRX098263) Global profiling of miRNA and the hairpin pre. (mixture) | SRR039154(GSM471931) HL1_siSrf2_smallRNAseq. (muscle) | SRR346415(SRX098256) source: Testis. (Testes) | SRR206939(GSM723280) other. (brain) | Ago3IP812(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) | SRR037921(GSM510459) e12p5_rep3. (embryo) | Ago1IP812(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | SRR116846(GSM678422) Small RNA deep sequencing from mouse epididym. (epididymis) | SRR038745(GSM527280) small RNA-Seq. (dgcr8 brain) | mjTestesWT2() Testes Data. (testes) | SRR553583(SRX182789) source: Cerebellum. (Cerebellum) | SRR042478(GSM539870) mouse lung tissue [09-002]. (lung) | SRR039153(GSM471930) HL1_siSrf1_smallRNAseq. (muscle) | SRR023852(GSM307161) ZHBT-c424hsmallrna_rep1. (cell line) | GSM510452(GSM510452) newborn_rep8. (total RNA) | SRR039152(GSM471929) HL1_siNon_smallRNAseq. (muscle) | SRR206942(GSM723283) other. (brain) | SRR038743(GSM527278) small RNA-Seq. (dicer brain) | SRR345203(SRX097264) source: size fractionated RNA from mouse hipp. (brain) | GSM509276(GSM509276) small RNA cloning by length. (testes) | SRR042480(GSM539872) mouse kidney tissue [09-002]. (kidney) | Ago1IP517(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................................................................................................................................................................................................................................................................TCAGGGGCTCTGGCCAGACTTG......................................................................................... | 22 | 1 | 57.00 | 57.00 | 13.00 | 10.00 | 6.00 | 10.00 | - | - | - | 2.00 | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | 1.00 | 1.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................................................................................................................................................TCAGGGGCTCTGGCCAGACTTGA........................................................................................ | 23 | 1 | 22.00 | 57.00 | 2.00 | 3.00 | 1.00 | - | 3.00 | - | - | - | 1.00 | - | 2.00 | - | - | 1.00 | 1.00 | - | - | - | - | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..............................................................................................................................................................................................................................................................................................................................................................................................CAGGGGCTCTGGCCAGACTTG......................................................................................... | 21 | 1 | 9.00 | 9.00 | 2.00 | 3.00 | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................................................................................................................................................................................CAGGGGCTCTGGCCAGACTTGA........................................................................................ | 22 | 1 | 7.00 | 9.00 | 3.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................................................................................................................................................................................................................................AAGACTGGCCTTTGTGTCTCA................................................... | 21 | 1 | 6.00 | 6.00 | - | - | - | - | 5.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................................................................................................................................................TCAGGGGCTCTGGCCAGACTTGT........................................................................................ | 23 | 1 | 5.00 | 57.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................................................................................................................................................................................................................................................................TACTTTGGGCCCGGCTGCGAGCAGA........... | 25 | 1 | 5.00 | 5.00 | - | - | - | - | - | - | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................................................................................................................................................TCAGGGGCTCTGGCCAGACT........................................................................................... | 20 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ....................................................................................................................................................................................................................................................................................................................................................................................................................................AAGACTGGCCTTTGTGTCTCAG.................................................. | 22 | 1 | 4.00 | 4.00 | - | - | - | - | - | 3.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................................................................................................................................................................................................................................GCAAGACTGGCCTTTGTGTCTCAG.................................................. | 24 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................................................................................................................................................TCAGGGGCTCTGGCCAGACTT.......................................................................................... | 21 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..............................................................................................................................................................................................................................................................................................................................................................................................CAGGGGCTCTGGCCAGACTTGG........................................................................................ | 22 | 1 | 3.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .......................ACAACCTTGAAACAGGAGATACTT............................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 24 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................................................................................................................................................................................CAGGGGCTCTGGCCAGACTTGT........................................................................................ | 22 | 1 | 2.00 | 9.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................................................................ATCCTACATGGATCCAAA.................................................................................................................................................................................................................... | 18 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................................................................................................................................................................................................................................................................................................................................TGGGCCCGGCTGCGAGCAGA........... | 20 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................................................................GGATCCTTAAGACAAGGT........................................................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................................................................................................................................................................................................................................ACTGGCCTTTGTGTCAGAG.................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................CGGGAGGAGAGGAACATC............................................................................................................................................................................................................................................................................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................AGGAGAGGAACGGGGGA.......................................................................................................................................................................................................................................................................................................................... | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................................................................................................................................................................................................................................................................................................................ACTGGCCTTTGTGTCTCCCA................................................. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................................................................................................................................................CTGGGACCCTTACCTCCAGGC...................................................................................................................................................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................................................................................................................................................................................................................................................................AGGGGCTCTGGCCAGACTTGG........................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................AGATTTGCGGATGG..................................................................................................................................................................................................................................................................................... | 14 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................ACAACCTTGAAACAGGAGAT................................................................................................................................................................................................................................................................................................................................................................................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................................................................................................................................................TCAGGGGCTCTGGCCAGACTTAAT....................................................................................... | 24 | 1 | 1.00 | 3.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................................................................................................................ATCTGGGGAGAGGGTGT........................................................................................................................................................................ | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................................................................................................................................................................................................................................................TCAGGGGCTCTGGCCAGACTTGAA....................................................................................... | 24 | 1 | 1.00 | 57.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................................................................................................................................................TCAGGGGCTCTGGCCAGACTTGGTGTC.................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...........................................................................................................................................................CCCGGGAGGAGAGGAAGTC.............................................................................................................................................................................................................................................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................................................................................................................................................................................................................................................TCAGGGGCTCTGGCCAGACTTGAAA...................................................................................... | 25 | 1 | 1.00 | 57.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................................................................................................................................................TCAGGGGCTCTGGCCAGACTTTA........................................................................................ | 23 | 1 | 1.00 | 3.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................................................................................................................................................................TCAGGGGCTCTGGCCAGACTTT......................................................................................... | 22 | 1 | 1.00 | 3.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................................................................................................................................................................................................................................AGACTGGCCTTTGTGTCTCAGT................................................. | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | |
| ....................................................................................................................................................................AGAGGAACGGGGTCCAGGT..................................................................................................................................................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................................................................................................................................................................................................................................................................................................................................................................ACTTTGGGCCCGGCTGCGAGCAGAAG......... | 26 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................AAGATTTGCGGATGGTTGC................................................................................................................................................................................................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................................CCCGGGAGGAGAGGAATG............................................................................................................................................................................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | |
| ...............................................................................................................................................ACACTGTCCCAGCCCGGGAGGAGAGGAACGGGGTCCTTAGGCTCCCTGTATGCTTCCAAGATTTGCGGATGGGCTGTGAGTGGGTCAGGG................................................................................................................................................................................................................................................................... | 90 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................................................................................................................................................................AGGAGCTTCTTTGTCAGGGGCTCTG................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................................................................................................................................................................................CAGGGGCTCTGGCCAGACTTGGAA...................................................................................... | 24 | 1 | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................................................................................................................................................................................................................................GCCTTTGTGTCTCAGCTTC.............................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TGCCCAGCAGGAGCAATGTTTACACAACCTTGAAACAGGAGATACTTAGAGTAAGCATTACAGTGTCACATCCATATTGCACCAAGATATAGGGGTGGAGAAACCAGCATTTGAATGATACAAGTGCACAACACATTCTGCCTACACTGTCCCAGCCCGGGAGGAGAGGAACGGGGTCCTTAGGCTCCCTGTATGCTTCCAAGATTTGCGGATGGGCTGTGAGTGGGTCAGGGTAGCTGGCCTTAGAGACAGCCCTAGTGAGATCCTACATGGATCCTTAAGACAAGGTCTGGGACCCTTACCTCCCATCTGGGGAGAGGGTAGGAGGAGTGTAGGGTAAGGTCAGCTGTCCCCACTGTTGAGAGTGGAGGAGCTTCTTTGTCAGGGGCTCTGGCCAGACTTGGTGTCCTAATCTGATGCAAGACTGGCCTTTGTGTCTCAGGGTGTCGCCAAGGCTACTTTGGGCCCGGCTGCGAGCAGAAGTGCAGGTGT ................................................................................................................................................................................................................................................................................................................................................................................................(((((...((((((.(((.(((((.......)))))))).)))))).....))))).................................................... ........................................................................................................................................................................................................................................................................................................................................................................................377..............................................................442................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | mjLiverWT1() Liver Data. (liver) | SRR345204(SRX097265) source: size fractionated RNA from mouse hipp. (brain) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | mjTestesWT4() Testes Data. (testes) | SRR685339(GSM1079783) Small RNAs (15-50 nts in length) from immorta. (cell line) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR306536(GSM750579) 19-24nt. (ago2 brain) | SRR279904(GSM689054) cell type: mouse embryonic stem cellcell line. (ESC) | SRR346419(SRX098260) Global profiling of miRNA and the hairpin pre. (Lung) | SRR206940(GSM723281) other. (brain) | SRR037937(GSM510475) 293cand2. (cell line) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR345201(SRX097262) source: size fractionated RNA from mouse hipp. (brain) | GSM510456(GSM510456) newborn_rep12. (total RNA) | SRR346423(SRX098265) Global profiling of miRNA and the hairpin pre. (Brain) | SRR037914(GSM510451) newborn_rep7. (total RNA) | RuiDGCR8KO(Rui) DGCR8-dependent microRNA biogenesis is essent. (dgcr8 skin) | SRR345199(SRX097260) source: size fractionated RNA from mouse hipp. (brain) | SRR206941(GSM723282) other. (brain) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) | SRR073954(GSM629280) total RNA. (blood) | SRR065050(SRR065050) Tissue-specific Regulation of Mouse MicroRNA . (liver) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | GSM510455(GSM510455) newborn_rep11. (total RNA) | SRR345206(SRX097267) source: size fractionated RNA from mouse hipp. (brain) | SRR037905(GSM510441) brain_rep2. (brain) | GSM510454(GSM510454) newborn_rep10. (total RNA) | SRR073955(GSM629281) total RNA. (blood) | SRR042486(GSM539878) mouse ovaries [09-002]. (ovary) | SRR346421(SRX098263) Global profiling of miRNA and the hairpin pre. (mixture) | SRR039154(GSM471931) HL1_siSrf2_smallRNAseq. (muscle) | SRR346415(SRX098256) source: Testis. (Testes) | SRR206939(GSM723280) other. (brain) | Ago3IP812(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) | SRR037921(GSM510459) e12p5_rep3. (embryo) | Ago1IP812(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | SRR116846(GSM678422) Small RNA deep sequencing from mouse epididym. (epididymis) | SRR038745(GSM527280) small RNA-Seq. (dgcr8 brain) | mjTestesWT2() Testes Data. (testes) | SRR553583(SRX182789) source: Cerebellum. (Cerebellum) | SRR042478(GSM539870) mouse lung tissue [09-002]. (lung) | SRR039153(GSM471930) HL1_siSrf1_smallRNAseq. (muscle) | SRR023852(GSM307161) ZHBT-c424hsmallrna_rep1. (cell line) | GSM510452(GSM510452) newborn_rep8. (total RNA) | SRR039152(GSM471929) HL1_siNon_smallRNAseq. (muscle) | SRR206942(GSM723283) other. (brain) | SRR038743(GSM527278) small RNA-Seq. (dicer brain) | SRR345203(SRX097264) source: size fractionated RNA from mouse hipp. (brain) | GSM509276(GSM509276) small RNA cloning by length. (testes) | SRR042480(GSM539872) mouse kidney tissue [09-002]. (kidney) | Ago1IP517(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................CTGTCCCAGCCCGGGACTGA...................................................................................................................................................................................................................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................................................GGCTCCCTGTATGCTGAA.................................................................................................................................................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................................................................................................................................................................................................................................................................TGGTGTCCTAATCTGATGCAAG..................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................CCCTGTATGCTTCCATCC................................................................................................................................................................................................................................................................................................ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................................................................................................................................................................................................................................GCTCTGGCCAGACTTG......................................................................................... | 16 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................CTGTCCCAGCCCGGGGGC........................................................................................................................................................................................................................................................................................................................................ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |