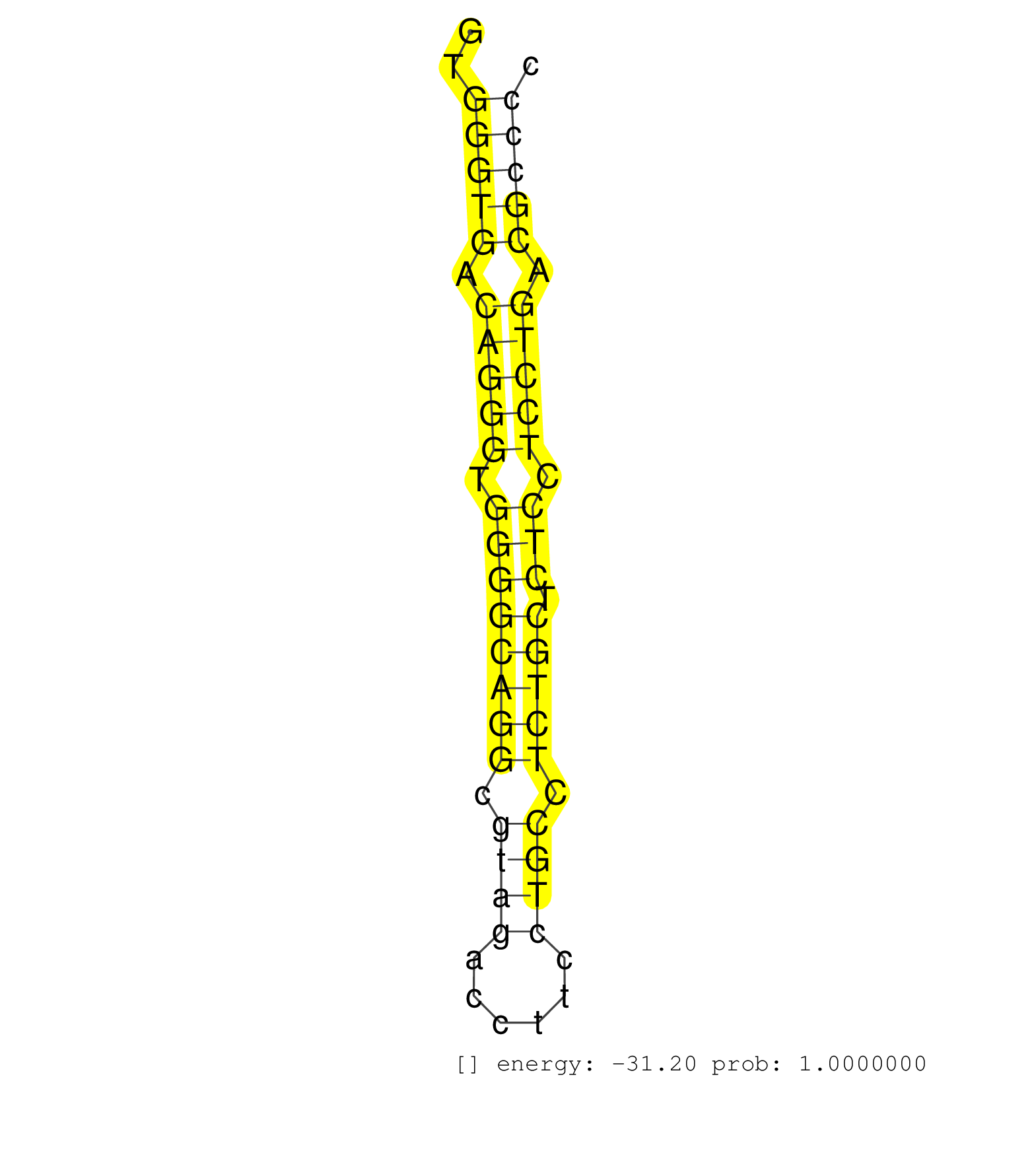
| (4) AGO1.ip | (7) AGO2.ip | (1) AGO3.ip | (4) B-CELL | (33) BRAIN | (3) CELL-LINE | (2) DCR.mut | (2) DGCR8.mut | (9) EMBRYO | (6) ESC | (1) FIBROBLAST | (2) KIDNEY | (2) LIVER | (1) LUNG | (2) LYMPH | (13) OTHER | (7) OTHER.mut | (3) PIWI.ip | (2) PIWI.mut | (4) SKIN | (4) SPLEEN | (23) TESTES | (2) THYMUS | (1) UTERUS |
| AGAAGCAGCTGATGGACAGGTCTACTGAACTGGAGCAGCAGCGACTGAGGGTGGGTGACAGGGTGGGGCAGGCGTAGACCTTCCTGCCTCTGCTCTCCTCCTGACGCCCCTCCTCCTCACCTAGGACACAGAACACAGCCAGGACCTGGACAGCGCTCTTCTGCGCCTAGAGGA ....................................................(((((.(((((.((((((((.((((......)))).))))).))).))))).)))))................................................................. ..................................................51.........................................................110.............................................................. | Size | Perfect hit | Total Norm | Perfect Norm | SRR546155(SRX180174) Global profiling of miRNA and the hairpin pre. (ago2 Neuroblastoma) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR038744(GSM527279) small RNA-Seq. (brain) | GSM314557(GSM314557) ESC dgcr8 (Illumina). (ESC) | SRR206940(GSM723281) other. (brain) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR345203(SRX097264) source: size fractionated RNA from mouse hipp. (brain) | SRR039610(GSM527274) small RNA-Seq. (brain) | SRR038741(GSM527276) small RNA-Seq. (brain) | SRR553582(SRX182788) source: Brain. (Brain) | SRR038745(GSM527280) small RNA-Seq. (dgcr8 brain) | SRR345204(SRX097265) source: size fractionated RNA from mouse hipp. (brain) | SRR553583(SRX182789) source: Cerebellum. (Cerebellum) | mjTestesWT4() Testes Data. (testes) | SRR402760(SRX117943) source: ES E14 male cells. (ESC) | SRR553585(SRX182791) source: Kidney. (Kidney) | SRR346424(SRX098266) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR038743(GSM527278) small RNA-Seq. (dicer brain) | SRR206939(GSM723280) other. (brain) | mjTestesKO6() Testes Data. (Zcchc11 testes) | Ago1IP517(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | SRR060845(GSM561991) total RNA. (brain) | SRR206941(GSM723282) other. (brain) | SRR346417(SRX098258) Global profiling of miRNA and the hairpin pre. (Brain) | SRR279906(GSM689056) cell type: mouse embryonic stem cellcell line. (ESC) | SRR023847(GSM307156) mESv6.5smallrna_rep1. (cell line) | SRR306534(GSM750577) 19-24nt. (ago2 brain) | SRR306533(GSM750576) 19-24nt. (ago2 brain) | Ago2IP517(Rui) Quantitative functions of Argonaute proteins . (ago2 skin) | SRR346425(SRX098267) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR037920(GSM510458) e12p5_rep2. (embryo) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR279907(GSM689057) cell type: mouse embryonic stem cellcell line. (ESC) | SRR037921(GSM510459) e12p5_rep3. (embryo) | GSM510444(GSM510444) brain_rep5. (brain) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR346421(SRX098263) Global profiling of miRNA and the hairpin pre. (mixture) | SRR346423(SRX098265) Global profiling of miRNA and the hairpin pre. (Brain) | SRR206942(GSM723283) other. (brain) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | Ago1IP812(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR116846(GSM678422) Small RNA deep sequencing from mouse epididym. (epididymis) | GSM314552(GSM314552) ESC wild type (Illumina). (ESC) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR042469(GSM539861) mouse Th2 cells [09-002]. (lymph) | SRR345196(SRX097257) source: size fractionated RNA from mouse hipp. (brain) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR345207(SRX097268) source: size fractionated RNA from mouse hipp. (brain) | SRR042480(GSM539872) mouse kidney tissue [09-002]. (kidney) | SRR059766(GSM562828) DN3_Drosha. (thymus) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR345205(SRX097266) source: size fractionated RNA from mouse hipp. (brain) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR042464(GSM539856) mouse CD4+CD8+ T cells (thymus) [09-002]. (thymus) | SRR391847(GSM852142) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR039153(GSM471930) HL1_siSrf1_smallRNAseq. (muscle) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjLiverKO1() Liver Data. (Zcchc11 liver) | SRR042471(GSM539863) mouse TFHS cells [09-002]. (spleen) | SRR345202(SRX097263) source: size fractionated RNA from mouse hipp. (brain) | SRR042443(GSM539835) mouse pro B cells [09-002]. (b cell) | SRR059772(GSM562834) CD4_Drosha. (spleen) | SRR042467(GSM539859) mouse T cells activated in vitro with Concanavalin A [09-002]. (spleen) | SRR039183(GSM485323) shLuc.1309 (-dox). (fibroblast) | SRR346419(SRX098260) Global profiling of miRNA and the hairpin pre. (Lung) | SRR685340(GSM1079784) Small RNAs (15-50 nts in length) from immorta. (dicer cell line) | SRR042452(GSM539844) mouse B1 B cells [09-002]. (b cell) | SRR553586(SRX182792) source: Testis. (testes) | SRR306539(GSM750582) 19-24nt. (ago2 brain) | SRR525241(SRA056111/SRX170317) Mus musculus domesticus miRNA sequencing. (ago1 Blood) | SRR039154(GSM471931) HL1_siSrf2_smallRNAseq. (muscle) | SRR042453(GSM539845) mouse marginal zone B cells (spleen) [09-002]. (b cell) | SRR553603(SRX182809) source: Spermatids. (Spermatids) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR525240(SRA056111/SRX170316) Mus musculus domesticus miRNA sequencing. (ago1 Blood) | SRR023850(GSM307159) ZHBT-c40hsmallrna_rep1. (cell line) | SRR306532(GSM750575) 19-24nt. (ago2 brain) | SRR042474(GSM539866) mouse embryonic stem cells [09-002]. (ESC) | SRR059769(GSM562831) Treg_Drosha. (spleen) | GSM361399(GSM361399) CGNP_P6_p53--_Ink4c--_rep2. (brain) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR346420(SRX098261) Global profiling of miRNA and the hairpin pre. (Uterus) | SRR037906(GSM510442) brain_rep3. (brain) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR553602(SRX182808) source: Sertoli cells. (Sertoli cells) | mjTestesWT2() Testes Data. (testes) | SRR042470(GSM539862) mouse Th17 cells [09-002]. (lymph) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR345199(SRX097260) source: size fractionated RNA from mouse hipp. (brain) | SRR306536(GSM750579) 19-24nt. (ago2 brain) | SRR346422(SRX098264) Global profiling of miRNA and the hairpin pre. (Muscle) | SRR042447(GSM539839) mouse mature B cells (spleen) replicate 2 [09-002]. (b cell) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | GSM361394(GSM361394) CGNP_P6_wt_rep1. (brain) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | Ago3IP805(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................TGCCTCTGCTCTCCTCCTGACG.................................................................... | 22 | 1 | 139.00 | 139.00 | 21.00 | 13.00 | 13.00 | 4.00 | 8.00 | 2.00 | 7.00 | 6.00 | 7.00 | 4.00 | 6.00 | 5.00 | 3.00 | - | - | 2.00 | - | 1.00 | 1.00 | 2.00 | - | - | 3.00 | 1.00 | 2.00 | - | 4.00 | - | - | - | - | - | 1.00 | - | 2.00 | 1.00 | - | - | 2.00 | 2.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | 2.00 | 1.00 | - | - | 2.00 | 1.00 | - | 1.00 | - | 1.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - |
| ....................................................................................TGCCTCTGCTCTCCTCCTGACGC................................................................... | 23 | 1 | 72.00 | 72.00 | 45.00 | 3.00 | - | 2.00 | 2.00 | 4.00 | - | - | - | - | 1.00 | - | - | - | - | - | 2.00 | 1.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TGCCTCTGCTCTCCTCCTGACGT................................................................... | 23 | 1 | 28.00 | 139.00 | 6.00 | 1.00 | 1.00 | 2.00 | - | - | - | - | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - |
| ....................................................................................TGCCTCTGCTCTCCTCCTGAC..................................................................... | 21 | 1 | 17.00 | 17.00 | 4.00 | 3.00 | - | 1.00 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TGCCTCTGCTCTCCTCCTGA...................................................................... | 20 | 1 | 12.00 | 12.00 | 5.00 | 2.00 | - | 1.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TGCCTCTGCTCTCCTCCTGACGA................................................................... | 23 | 1 | 12.00 | 139.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TGCCTCTGCTCTCCTCCTGACA.................................................................... | 22 | 1 | 10.00 | 17.00 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................TCTGCTCTCCTCCTGACG.................................................................... | 18 | 1 | 9.00 | 9.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TGCCTCTGCTCTCCTCCTGACT.................................................................... | 22 | 1 | 7.00 | 17.00 | 2.00 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................CACAGCCAGGACCTGGACAGC.................... | 21 | 1 | 6.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................AGCAGCAGCGACTGAGG............................................................................................................................ | 17 | 1 | 6.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TGCCTCTGCTCTCCTCCTGGT..................................................................... | 21 | 2 | 6.00 | 1.50 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................TGAACTGGAGCAGCAGCG................................................................................................................................... | 18 | 1 | 6.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TGCCTCTGCTCTCCTCCTGTGT.................................................................... | 22 | 2 | 5.00 | 1.50 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................GCGTAGACCTTCCTGCC...................................................................................... | 17 | 1 | 4.00 | 4.00 | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................TCTGCTCTCCTCCTGACGCT.................................................................. | 20 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TGCCTCTGCTCTCCTCCTGAA..................................................................... | 21 | 1 | 4.00 | 12.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TGCCTCTGCTCTCCTCCTGACGCT.................................................................. | 24 | 1 | 4.00 | 72.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................TCTGCTCTCCTCCTGACGC................................................................... | 19 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................ACTGAACTGGAGCAGCAGCGACT................................................................................................................................ | 23 | 1 | 3.00 | 3.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TGCCTCTGCTCTCCTCCTGACGTA.................................................................. | 24 | 1 | 3.00 | 139.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................TCTGCTCTCCTCCTGACGCCCCT............................................................... | 23 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................TCTGCTCTCCTCCTGACGCC.................................................................. | 20 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TGCCTCTGCTCTCCTCCTCG...................................................................... | 20 | 3.00 | 0.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................TCTGCTCTCCTCCTGACGCCTTTT.............................................................. | 24 | 1 | 3.00 | 3.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................CTCTGCTCTCCTCCTGACG.................................................................... | 19 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................CGCCCCTCCTCCTCACCTAGT................................................. | 21 | 3.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................GAACTGGAGCAGCAGCGACT................................................................................................................................ | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TGCCTCTGCTCTCCTCCTGACGCA.................................................................. | 24 | 1 | 2.00 | 72.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................CTCTGCTCTCCTCCTGACGCC.................................................................. | 21 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TGCCTCTGCTCTCCTCCTGAAA.................................................................... | 22 | 1 | 2.00 | 12.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TGCCTCTGCTCTCCTCCTGACGCAT................................................................. | 25 | 1 | 2.00 | 72.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................CCTTCCTGCCTCTGCGAT.............................................................................. | 18 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | |
| ........................................................................................TCTGCTCTCCTCCTGACGA................................................................... | 19 | 1 | 2.00 | 9.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................CTACTGAACTGGAGCAGCAGCGACTG............................................................................................................................... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGGTGACAGGGTGGGGCAGG...................................................................................................... | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................CGCCCCTCCTCCTCACCTAGA................................................. | 21 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | |
| ....................................................................................TGCCTCTGCTCTCCTCCTGAGT.................................................................... | 22 | 1 | 2.00 | 12.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGGTGACAGGGTGGGGCAGGCGTTA................................................................................................. | 27 | 1 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGGTGACAGGGTGGGGCAGGCGTAGACCTTCCTGCCTCTGCTCTCCTCCTGACGT................................................................... | 57 | 1 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TGCCTCTGCTCTCCTCCTG....................................................................... | 19 | 2 | 1.50 | 1.50 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................TGCTCTCCTCCTGACGCCCCTCA............................................................. | 23 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................TGCCTCTGCTCTCCTCCTCCGC.................................................................... | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................................GCCAGGACCTGGACAGC.................... | 17 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................TGCTCTCCTCCTGACGCCCCTTT............................................................. | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....GCAGCTGATGGACAGGTCTACTGA.................................................................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................CTCTGCTCTCCTCCTGACGC................................................................... | 20 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................TCTGCTCTCCTCCTGACGCCT................................................................. | 21 | 1 | 1.00 | 3.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGGTGACAGGGTGGGGCA........................................................................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................TACTGAACTGGAGCAGCAGCGACTGAGGGACAC....................................................................................................................... | 33 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................GGACACAGAACACAGCCAGGACCTGGA........................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............TGGACAGGTCTACTGAACT............................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............TGGACAGGTCTACTGAACTGGAGCAG........................................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................GGGTGACAGGGTGGGGCAGG...................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................TCTGCTCTCCTCCTGACGCCAATA.............................................................. | 24 | 1 | 1.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................TTCCTGCCTCTGCTCTCCTCCTGAC..................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGGTGACAGGGTGGGGCAGGA..................................................................................................... | 23 | 1 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ......................TACTGAACTGGAGCAGCAGCGACTAA.............................................................................................................................. | 26 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................AGGGTGGGGCAGGCG.................................................................................................... | 15 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................TCTGCTCTCCTCCTGACGAA.................................................................. | 20 | 1 | 1.00 | 9.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................AGGTCTACTGAACTGGAGCAGCAGCGACTGAGG............................................................................................................................ | 33 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................GTCTACTGAACTGGAGCAGCAGCGACT................................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................CACAGAACACAGCCAGGACCTGGACAG..................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TGCCTCTGCTCTCCTCCTGACGCAG................................................................. | 25 | 1 | 1.00 | 72.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................TGCTCTCCTCCTGACGCCCCTCAT............................................................ | 24 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................TGACAGGGTGGGGCAGGCGTAG................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................TACTGAACTGGAGCAGCAGCGACTGA.............................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................AGGGTGGGGCAGGCGTAGA................................................................................................ | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................TCCTCCTGACGCCCCTCCTCCTCACCTAGT................................................. | 30 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................TGGGGCAGGCGTAGAA............................................................................................... | 16 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................TGCCTCTGCTCTCCTCCTGT...................................................................... | 20 | 2 | 1.00 | 1.50 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................CTCTGCTCTCCTCCTGACGT................................................................... | 20 | 1 | 1.00 | 3.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................CAGGGTGGGGCAGGCAATT................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................GGTGACAGGGTGGGGCAGGC..................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGGTGACAGGGTGGGGCAGGCGT................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................GTGACAGGGTGGGGCAGGCGT................................................................................................... | 21 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGGTGACAGGGTGGGG.......................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TGCCTCTGCTCTCCTCCTGCG..................................................................... | 21 | 2 | 1.00 | 1.50 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................TGAACTGGAGCAGCAGCGACTGAGGGA.......................................................................................................................... | 27 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................ACAGAACACAGCCAGGACCTGGAC....................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................ACTGAACTGGAGCAGCAGCGAC................................................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TGCCTCTGCTCTCCTCCTTAAA.................................................................... | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................TGCCTCTGCTCTCCTCCTGCGC.................................................................... | 22 | 2 | 1.00 | 1.50 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TGCCTCTGCTCTCCTCCTGATT.................................................................... | 22 | 1 | 1.00 | 12.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................TCTGCTCTCCTCCTGACGCCCTT............................................................... | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................TCTGCTCTCCTCCTGACGCTCTT............................................................... | 23 | 1 | 1.00 | 4.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................GACACAGAACACAGCCAGGACCTGGACAGCGC.................. | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TGCCTCTGCTCTCCTCCTCCG..................................................................... | 21 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................CTGCCTCTGCTCTCCTCCTGCG..................................................................... | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................GAACTGGAGCAGCAGAAG.................................................................................................................................. | 18 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................TGCCTCTGCTCTCCTCCTGACGCTA................................................................. | 25 | 1 | 1.00 | 72.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TGCCTCTGCTCTCCTCCTGACGTT.................................................................. | 24 | 1 | 1.00 | 139.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................CTGCCTCTGCTCTCCTCCTGT...................................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................CACAGCCAGGACCTGGACAGCGCTCTTC............. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TGCCTCTGCTCTCCTCCTGACGGT.................................................................. | 24 | 1 | 1.00 | 139.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................TCTACTGAACTGGAGCAGCA...................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................ACTGGAGCAGCAGCGAC................................................................................................................................. | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................TACTGAACTGGAGCAGCAGCGACTG............................................................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................TAGACCTTCCTGCCTCTGCTCTC............................................................................. | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................ACTGAACTGGAGCAGCAGCGACTGAGG............................................................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................TCTGCTCTCCTCCTGACGCCTT................................................................ | 22 | 1 | 1.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................GGTGACAGGGTGGGGCAGGCGTAGA................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................TGGGGCAGGCGTAGAATTG............................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................CTGAACTGGAGCAGCAGCGACTGAGG............................................................................................................................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................GTGACAGGGTGGGGCAGGCGTAGA................................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................TACTGAACTGGAGCAGCAGCGACTGAGG............................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................TGACAGGGTGGGGCAGGCGGAGA................................................................................................ | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................AGCAGCAGCGACTGAGCG........................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | |
| ........................................................................................TCTGCTCTCCTCCTGACGCA.................................................................. | 20 | 1 | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TGCCTCTGCTCTCCTCCTGACGTAT................................................................. | 25 | 1 | 1.00 | 139.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................AGCAGCAGCGACTGAGGGAC......................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................TGCTCTCCTCCTGACGCCCCTCC............................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................GCCTCTGCTCTCCTCCTGACG.................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGGTGACAGGGTGGGGCAGGCGTAGACCTTCCTGCCTCTGCTCTCCTCCTGACGCCCCTCCTCCT......................................................... | 67 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGGTGACAGGGTGGGGCAGGCGTA.................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................GCGTAGACCTTCCTGCCTCT................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGGTGACAGGGTGGGGCAGGCGTC.................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGGGTGACAGGGTGGGGCAGGCGTAGACCTTCCTGCCTCTGCTCTCCTCCTGACG.................................................................... | 56 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................ACAGAACACAGCCAGGACC............................ | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................CTGCCTCTGCTCTCCTCCTGACGT................................................................... | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................CTGAACTGGAGCAGCAGCGACTGAGGG........................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................ACTGAACTGGAGCAGCAGC.................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................GCCTCTGCTCTCCTCCTGACGCT.................................................................. | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................GAACACAGCCAGGACCTGGACAGCG................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................GTAGACCTTCCTGCC...................................................................................... | 15 | 3 | 0.67 | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................ACTGAACTGGAGCAGC....................................................................................................................................... | 16 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| ....................................................................................TGCCTCTGCTCTCCTCCCTGT..................................................................... | 21 | 7 | 0.14 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TGCCTCTGCTCTCCTCC......................................................................... | 17 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TGCCTCTGCTCTCCTCCCGAC..................................................................... | 21 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................TGCCTCTGCTCTCCTCCCG....................................................................... | 19 | 7 | 0.14 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AGAAGCAGCTGATGGACAGGTCTACTGAACTGGAGCAGCAGCGACTGAGGGTGGGTGACAGGGTGGGGCAGGCGTAGACCTTCCTGCCTCTGCTCTCCTCCTGACGCCCCTCCTCCTCACCTAGGACACAGAACACAGCCAGGACCTGGACAGCGCTCTTCTGCGCCTAGAGGA ....................................................(((((.(((((.((((((((.((((......)))).))))).))).))))).)))))................................................................. ..................................................51.........................................................110.............................................................. | Size | Perfect hit | Total Norm | Perfect Norm | SRR546155(SRX180174) Global profiling of miRNA and the hairpin pre. (ago2 Neuroblastoma) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR038744(GSM527279) small RNA-Seq. (brain) | GSM314557(GSM314557) ESC dgcr8 (Illumina). (ESC) | SRR206940(GSM723281) other. (brain) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR345203(SRX097264) source: size fractionated RNA from mouse hipp. (brain) | SRR039610(GSM527274) small RNA-Seq. (brain) | SRR038741(GSM527276) small RNA-Seq. (brain) | SRR553582(SRX182788) source: Brain. (Brain) | SRR038745(GSM527280) small RNA-Seq. (dgcr8 brain) | SRR345204(SRX097265) source: size fractionated RNA from mouse hipp. (brain) | SRR553583(SRX182789) source: Cerebellum. (Cerebellum) | mjTestesWT4() Testes Data. (testes) | SRR402760(SRX117943) source: ES E14 male cells. (ESC) | SRR553585(SRX182791) source: Kidney. (Kidney) | SRR346424(SRX098266) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR038743(GSM527278) small RNA-Seq. (dicer brain) | SRR206939(GSM723280) other. (brain) | mjTestesKO6() Testes Data. (Zcchc11 testes) | Ago1IP517(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | SRR060845(GSM561991) total RNA. (brain) | SRR206941(GSM723282) other. (brain) | SRR346417(SRX098258) Global profiling of miRNA and the hairpin pre. (Brain) | SRR279906(GSM689056) cell type: mouse embryonic stem cellcell line. (ESC) | SRR023847(GSM307156) mESv6.5smallrna_rep1. (cell line) | SRR306534(GSM750577) 19-24nt. (ago2 brain) | SRR306533(GSM750576) 19-24nt. (ago2 brain) | Ago2IP517(Rui) Quantitative functions of Argonaute proteins . (ago2 skin) | SRR346425(SRX098267) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR037920(GSM510458) e12p5_rep2. (embryo) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR279907(GSM689057) cell type: mouse embryonic stem cellcell line. (ESC) | SRR037921(GSM510459) e12p5_rep3. (embryo) | GSM510444(GSM510444) brain_rep5. (brain) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR346421(SRX098263) Global profiling of miRNA and the hairpin pre. (mixture) | SRR346423(SRX098265) Global profiling of miRNA and the hairpin pre. (Brain) | SRR206942(GSM723283) other. (brain) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | Ago1IP812(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR116846(GSM678422) Small RNA deep sequencing from mouse epididym. (epididymis) | GSM314552(GSM314552) ESC wild type (Illumina). (ESC) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR042469(GSM539861) mouse Th2 cells [09-002]. (lymph) | SRR345196(SRX097257) source: size fractionated RNA from mouse hipp. (brain) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR345207(SRX097268) source: size fractionated RNA from mouse hipp. (brain) | SRR042480(GSM539872) mouse kidney tissue [09-002]. (kidney) | SRR059766(GSM562828) DN3_Drosha. (thymus) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR345205(SRX097266) source: size fractionated RNA from mouse hipp. (brain) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR042464(GSM539856) mouse CD4+CD8+ T cells (thymus) [09-002]. (thymus) | SRR391847(GSM852142) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR039153(GSM471930) HL1_siSrf1_smallRNAseq. (muscle) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | mjTestesKO7() Testes Data. (Zcchc11 testes) | mjLiverKO1() Liver Data. (Zcchc11 liver) | SRR042471(GSM539863) mouse TFHS cells [09-002]. (spleen) | SRR345202(SRX097263) source: size fractionated RNA from mouse hipp. (brain) | SRR042443(GSM539835) mouse pro B cells [09-002]. (b cell) | SRR059772(GSM562834) CD4_Drosha. (spleen) | SRR042467(GSM539859) mouse T cells activated in vitro with Concanavalin A [09-002]. (spleen) | SRR039183(GSM485323) shLuc.1309 (-dox). (fibroblast) | SRR346419(SRX098260) Global profiling of miRNA and the hairpin pre. (Lung) | SRR685340(GSM1079784) Small RNAs (15-50 nts in length) from immorta. (dicer cell line) | SRR042452(GSM539844) mouse B1 B cells [09-002]. (b cell) | SRR553586(SRX182792) source: Testis. (testes) | SRR306539(GSM750582) 19-24nt. (ago2 brain) | SRR525241(SRA056111/SRX170317) Mus musculus domesticus miRNA sequencing. (ago1 Blood) | SRR039154(GSM471931) HL1_siSrf2_smallRNAseq. (muscle) | SRR042453(GSM539845) mouse marginal zone B cells (spleen) [09-002]. (b cell) | SRR553603(SRX182809) source: Spermatids. (Spermatids) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR525240(SRA056111/SRX170316) Mus musculus domesticus miRNA sequencing. (ago1 Blood) | SRR023850(GSM307159) ZHBT-c40hsmallrna_rep1. (cell line) | SRR306532(GSM750575) 19-24nt. (ago2 brain) | SRR042474(GSM539866) mouse embryonic stem cells [09-002]. (ESC) | SRR059769(GSM562831) Treg_Drosha. (spleen) | GSM361399(GSM361399) CGNP_P6_p53--_Ink4c--_rep2. (brain) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR346420(SRX098261) Global profiling of miRNA and the hairpin pre. (Uterus) | SRR037906(GSM510442) brain_rep3. (brain) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR553602(SRX182808) source: Sertoli cells. (Sertoli cells) | mjTestesWT2() Testes Data. (testes) | SRR042470(GSM539862) mouse Th17 cells [09-002]. (lymph) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR345199(SRX097260) source: size fractionated RNA from mouse hipp. (brain) | SRR306536(GSM750579) 19-24nt. (ago2 brain) | SRR346422(SRX098264) Global profiling of miRNA and the hairpin pre. (Muscle) | SRR042447(GSM539839) mouse mature B cells (spleen) replicate 2 [09-002]. (b cell) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | GSM361394(GSM361394) CGNP_P6_wt_rep1. (brain) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | Ago3IP805(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................................ACAGCGCTCTTCTGCGTCTT..... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................CTCTGCTCTCCTCCTTTCA.................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | |
| ................................................................................TTCCTGCCTCTGCTCCTGA........................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................TCCTCCTCACCTAGGACACAGA.......................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................CCTCTGCTCTCCTCCTTTT..................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................CAGCGCTCTTCTGCG......... | 15 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |