| (12) AGO2.ip | (21) BRAIN | (1) DCR.mut | (1) DGCR8.mut | (11) EMBRYO | (3) ESC | (3) LIVER | (5) OTHER | (7) OTHER.mut | (4) PIWI.ip | (2) PIWI.mut | (2) SPLEEN | (19) TESTES | (2) TOTAL-RNA |
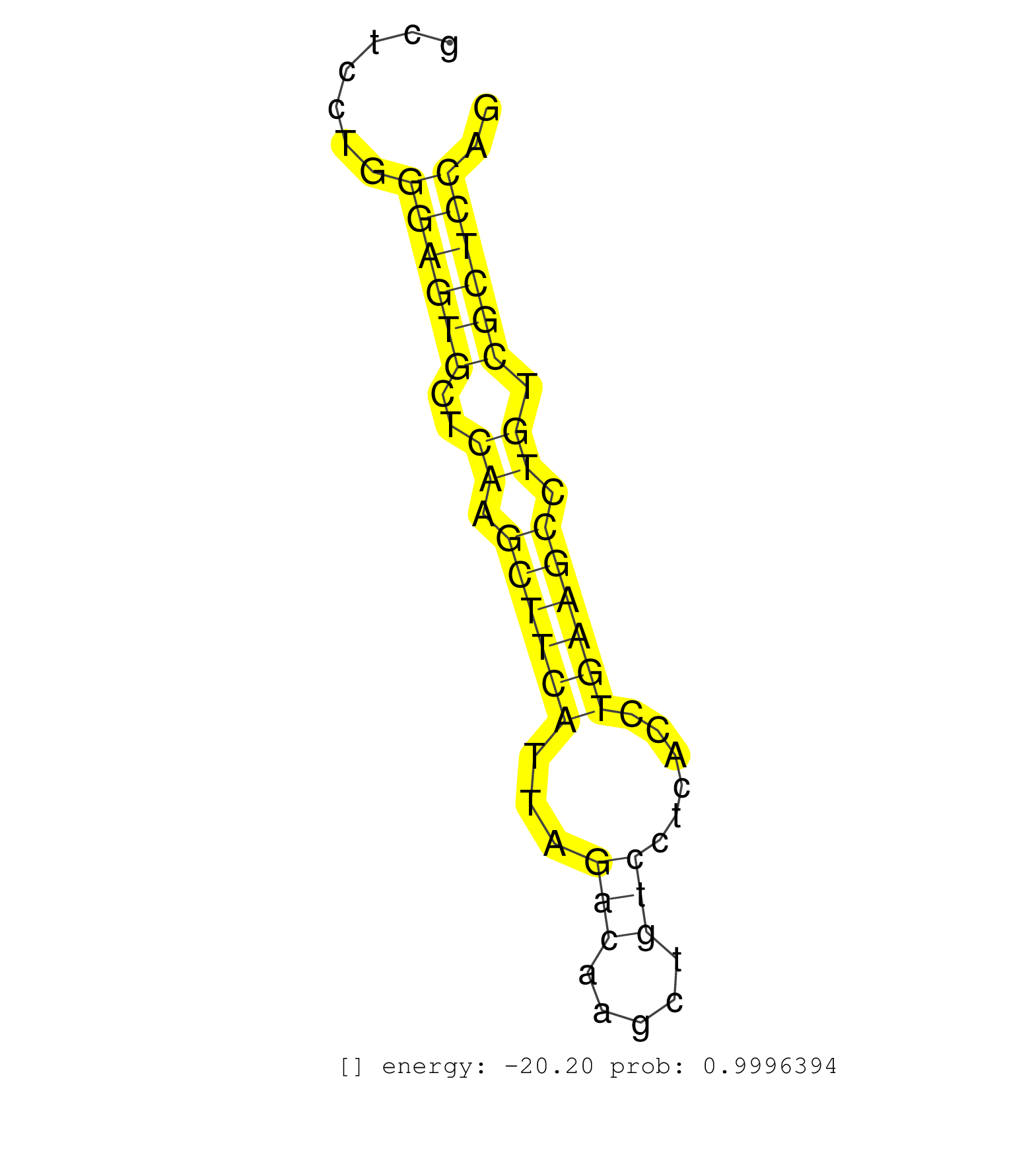
| CCTGCTGTGCCACCCGCCACCCATCAACCGCTCCTCACTCTGACTAGCAGGTAATCACGCCTCCTGCCCTCGCCAAGCCCTTCTTTTTCTTGTGTGGTCCATGAGGCGAATCCCACTGGGGTGAGGGCCTATGAAGCAGTAGGATGGGAGGCAGGGCATCTCGGTGCTCCTGGGAGTGCTCAAGCTTCATTAGACAAGCTGTCCTCACCTGAAGCCTGTCGCTCCAGGGCCCTGCTGACCCAGGTAGCTGTGGACGGAATGGCTGGTCCCCACAGAA ............................................................................................................................................................................((((((..((.((((((...(((.....)))......)))))).)).)))))).................................................... .....................................................................................................................................................................166..........................................................227................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR306535(GSM750578) 19-24nt. (ago2 brain) | SRR306536(GSM750579) 19-24nt. (ago2 brain) | SRR306542(GSM750585) 19-24nt. (ago2 brain) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR306537(GSM750580) 19-24nt. (ago2 brain) | SRR039153(GSM471930) HL1_siSrf1_smallRNAseq. (muscle) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | GSM640578(GSM640578) small RNA in the liver with paternal control. (liver) | SRR391847(GSM852142) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR037920(GSM510458) e12p5_rep2. (embryo) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR059773(GSM562835) CD4_Dicer. (spleen) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) | GSM640580(GSM640580) small RNA in the liver with paternal Low pro. (liver) | SRR279905(GSM689055) cell type: mouse embryonic stem cellcell line. (ESC) | SRR306539(GSM750582) 19-24nt. (ago2 brain) | GSM510454(GSM510454) newborn_rep10. (total RNA) | SRR345201(SRX097262) source: size fractionated RNA from mouse hipp. (brain) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR306529(GSM750572) 19-24nt. (ago2 brain) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR345203(SRX097264) source: size fractionated RNA from mouse hipp. (brain) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR306534(GSM750577) 19-24nt. (ago2 brain) | SRR038743(GSM527278) small RNA-Seq. (dicer brain) | SRR039152(GSM471929) HL1_siNon_smallRNAseq. (muscle) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR038744(GSM527279) small RNA-Seq. (brain) | SRR346421(SRX098263) Global profiling of miRNA and the hairpin pre. (mixture) | SRR306530(GSM750573) 19-24nt. (ago2 brain) | SRR346422(SRX098264) Global profiling of miRNA and the hairpin pre. (Muscle) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR306540(GSM750583) 19-24nt. (ago2 brain) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | GSM314557(GSM314557) ESC dgcr8 (Illumina). (ESC) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR059769(GSM562831) Treg_Drosha. (spleen) | SRR037912(GSM510449) newborn_rep5. (total RNA) | SRR306544(GSM750587) 19-24nt. (ago2 brain) | SRR038745(GSM527280) small RNA-Seq. (dgcr8 brain) | SRR037921(GSM510459) e12p5_rep3. (embryo) | SRR553602(SRX182808) source: Sertoli cells. (Sertoli cells) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM510444(GSM510444) brain_rep5. (brain) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR279903(GSM689053) cell type: mouse embryonic stem cellcell line. (ESC) | SRR306531(GSM750574) 19-24nt. (ago2 brain) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................................................................................ACCTGAAGCCTGTCGCTCCAGT................................................. | 22 | 1 | 30.00 | 4.00 | 5.00 | 5.00 | 2.00 | - | 3.00 | 4.00 | - | 1.00 | 1.00 | - | 1.00 | - | - | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...............................................................................................................................................................................................................CCTGAAGCCTGTCGCTCCAGT................................................. | 21 | 10.00 | 0.00 | 2.00 | 1.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | |
| ..........................................................................................................................................................................TGGGAGTGCTCAAGCTTCATTAG.................................................................................... | 23 | 1 | 8.00 | 8.00 | - | - | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..............................................................................................................................................................................................................ACCTGAAGCCTGTCGCTCCA................................................... | 20 | 1 | 5.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................ACCTGAAGCCTGTCGCTCCAG.................................................. | 21 | 1 | 4.00 | 4.00 | - | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................ACCTGAAGCCTGTCGCTCCAGA................................................. | 22 | 1 | 4.00 | 4.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................TGGGAGTGCTCAAGCTTCATT...................................................................................... | 21 | 1 | 3.00 | 3.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................TGGGAGTGCTCAAGCTTCATTA..................................................................................... | 22 | 1 | 3.00 | 3.00 | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................CCTCACTCTGACTAGCAGG.................................................................................................................................................................................................................................. | 19 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................TGGGAGTGCTCAAGCTTCATTAGT................................................................................... | 24 | 1 | 2.00 | 8.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................ACCTGAAGCCTGTCGCTC..................................................... | 18 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................................TGCTGACCCAGGTAGCTGTGGACGG.................... | 25 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................CCTGAAGCCTGTCGCTCCAGA................................................. | 21 | 2.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................CTCACTCTGACTAGCAG................................................................................................................................................................................................................................... | 17 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................................ACCCAGGTAGCTGTGGACGGAATGGC.............. | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................TGGGAGTGCTCAAGCTTCAT....................................................................................... | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................ACCTGAAGCCTGTCGCTCCAT.................................................. | 21 | 1 | 1.00 | 5.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................CCTGAAGCCTGTCGCTCCAGTAA............................................... | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................................................................................................................TGACCCAGGTAGCTGTGGACGGAA.................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ........................CAACCGCTCCTCACTCTGACTAGCAG................................................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .........................................................................................................................................................................CTGGGAGTGCTCAAGCTTCATT...................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................ATCACGCCTCCTGCCAAC.............................................................................................................................................................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................................................................................CCTGAAGCCTGTCGCTCCAGTAT............................................... | 23 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................................................................CTGAAGCCTGTCGCTCCAGT................................................. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................................................CCTGGGAGTGCTCAAGCTTCATT...................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................TCTCGGTGCTCCTGGGAGTGCTCAAGCTTCATTAGACAAGCTGTCCTCACCTGAAGCCTGTCGCTCCAG.................................................. | 69 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................TGAAGCAGTAGGATGCACG............................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................................................................TCCAGGGCCCTGCTGTGCG.................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................................................................................................AGGTAGCTGTGGACGCGG.................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................................................................................................AGGTAGCTGTGGACGGAATGG............... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................CCTGGGAGTGCTCAAGCTTCAT....................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...................................................................................................................................TGAAGCAGTAGGATGGGAGGTTTT.......................................................................................................................... | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................................................................................GTCCTCACCTGAAGCACCA.......................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................................................ACCTGAAGCCTGTCGCTCCAA.................................................. | 21 | 1 | 1.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................ACCTGAAGCCTGTCGCTCCATT................................................. | 22 | 1 | 1.00 | 5.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................CTCACCTGAAGCCTGTTTTC...................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................TGAAGCAGTAGGATGTGTC............................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................................................................................................GGCTGGTCCCCACAGAAT | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................................................ACCTGAAGCCTGTCGCTCCAGTAA............................................... | 24 | 1 | 1.00 | 4.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................TGAAGCAGTAGGATGTAT................................................................................................................................ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................................................................................................................TGACCCAGGTAGCTGTGGACGGAATGGC.............. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................ACCGCTCCTCACTCTGACTAGCAGGGCCC.............................................................................................................................................................................................................................. | 29 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................TGAAGCAGTAGGATGTCT................................................................................................................................ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................TGAAGCAGTAGGATGTTTG............................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................................................................................................................TGCTGACCCAGGTAGCTGTGGACGGA................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................ACCTGAAGCCTGTCGCTCC.................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................TCCTCACTCTGACTAGCAGGT................................................................................................................................................................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................CCTGAAGCCTGTCGCTC..................................................... | 17 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| CCTGCTGTGCCACCCGCCACCCATCAACCGCTCCTCACTCTGACTAGCAGGTAATCACGCCTCCTGCCCTCGCCAAGCCCTTCTTTTTCTTGTGTGGTCCATGAGGCGAATCCCACTGGGGTGAGGGCCTATGAAGCAGTAGGATGGGAGGCAGGGCATCTCGGTGCTCCTGGGAGTGCTCAAGCTTCATTAGACAAGCTGTCCTCACCTGAAGCCTGTCGCTCCAGGGCCCTGCTGACCCAGGTAGCTGTGGACGGAATGGCTGGTCCCCACAGAA ............................................................................................................................................................................((((((..((.((((((...(((.....)))......)))))).)).)))))).................................................... .....................................................................................................................................................................166..........................................................227................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR306535(GSM750578) 19-24nt. (ago2 brain) | SRR306536(GSM750579) 19-24nt. (ago2 brain) | SRR306542(GSM750585) 19-24nt. (ago2 brain) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR306537(GSM750580) 19-24nt. (ago2 brain) | SRR039153(GSM471930) HL1_siSrf1_smallRNAseq. (muscle) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | GSM640578(GSM640578) small RNA in the liver with paternal control. (liver) | SRR391847(GSM852142) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR037920(GSM510458) e12p5_rep2. (embryo) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR059773(GSM562835) CD4_Dicer. (spleen) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) | GSM640580(GSM640580) small RNA in the liver with paternal Low pro. (liver) | SRR279905(GSM689055) cell type: mouse embryonic stem cellcell line. (ESC) | SRR306539(GSM750582) 19-24nt. (ago2 brain) | GSM510454(GSM510454) newborn_rep10. (total RNA) | SRR345201(SRX097262) source: size fractionated RNA from mouse hipp. (brain) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR306529(GSM750572) 19-24nt. (ago2 brain) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR029040(GSM433292) 6w_hetero_tdrd6-KO. (tdrd6 testes) | SRR345203(SRX097264) source: size fractionated RNA from mouse hipp. (brain) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR306534(GSM750577) 19-24nt. (ago2 brain) | SRR038743(GSM527278) small RNA-Seq. (dicer brain) | SRR039152(GSM471929) HL1_siNon_smallRNAseq. (muscle) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | GSM475281(GSM475281) total RNA. (testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR038744(GSM527279) small RNA-Seq. (brain) | SRR346421(SRX098263) Global profiling of miRNA and the hairpin pre. (mixture) | SRR306530(GSM750573) 19-24nt. (ago2 brain) | SRR346422(SRX098264) Global profiling of miRNA and the hairpin pre. (Muscle) | SRR034120(GSM466730) Mili IP_Tdrd9-/- replicate1. (mili testes) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR306540(GSM750583) 19-24nt. (ago2 brain) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | GSM314557(GSM314557) ESC dgcr8 (Illumina). (ESC) | SRR034121(GSM466731) Mili IP_Tdrd9-/- replicate2. (mili testes) | SRR059769(GSM562831) Treg_Drosha. (spleen) | SRR037912(GSM510449) newborn_rep5. (total RNA) | SRR306544(GSM750587) 19-24nt. (ago2 brain) | SRR038745(GSM527280) small RNA-Seq. (dgcr8 brain) | SRR037921(GSM510459) e12p5_rep3. (embryo) | SRR553602(SRX182808) source: Sertoli cells. (Sertoli cells) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM510444(GSM510444) brain_rep5. (brain) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR279903(GSM689053) cell type: mouse embryonic stem cellcell line. (ESC) | SRR306531(GSM750574) 19-24nt. (ago2 brain) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................................................................GACAAGCTGTCCTCACCA................................................................... | 18 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................................................................................................................CTGCTGACCCAGGTATCTA........................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................................................................GCCCTGCTGACCCAGGTA............................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................................................................................................CGCTCCAGGGCCCTGCGTA....................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................................................AGCCTGTCGCTCCAG.................................................. | 15 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................CCTTCTTTTTCTTGTGTGGTCCATGA............................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................ACGCCTCCTGCCCTCCTG........................................................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................................................................GCCCTGCTGACCCAGGTAA.............................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........CCACCCGCCACCCATAGT.......................................................................................................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....CTGTGCCACCCGCCAGAG............................................................................................................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................................................................................................CTGTCGCTCCAGGGC............................................... | 15 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |