| (1) AGO.mut | (1) AGO1.ip | (9) AGO2.ip | (5) B-CELL | (21) BRAIN | (4) CELL-LINE | (1) DGCR8.mut | (5) EMBRYO | (2) ESC | (2) FIBROBLAST | (2) LIVER | (1) LYMPH | (9) OTHER | (4) OTHER.mut | (2) PIWI.ip | (2) SPLEEN | (16) TESTES | (2) THYMUS |
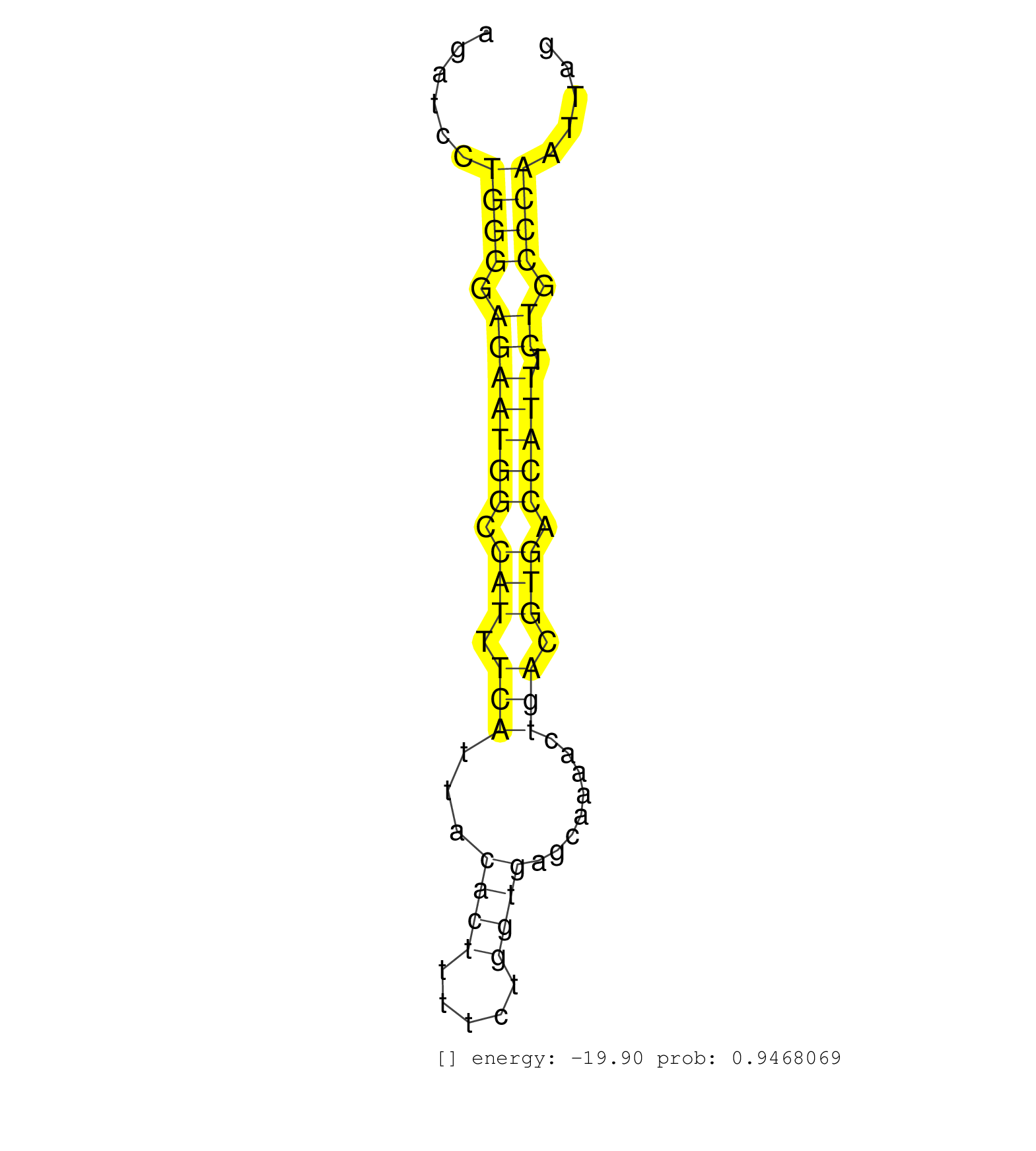
| CCCTTAAAGGAGGACAGATTTTGGAGTTTAAGTTTGCATGGGAATATTGGTGATCTGTCTCTGCTCTTGTGCTCAGTGCTTATGGGTGTGTACTTCAGGTGAAAGCACAGGCGTGTAGAAAAAAAGATCCTGGGGAGAATGGCCATTTCATTACACTTTTCTGGTGAGCAAAACTGACGTGACCATTTCTGCCCAATTAGGAGCTCTTGTCCCTGGCGAAACGGAAGCGCAGTGACTCCGAGGAGAAGGA ..................................................................................................................................((((.(((((((.(((.(((...((((.....))))........))).))).))))).)).))))....................................................... ............................................................................................................................125........................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR546155(SRX180174) Global profiling of miRNA and the hairpin pre. (ago2 Neuroblastoma) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR038744(GSM527279) small RNA-Seq. (brain) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR306537(GSM750580) 19-24nt. (ago2 brain) | mjLiverKO1() Liver Data. (Zcchc11 liver) | SRR553582(SRX182788) source: Brain. (Brain) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR042469(GSM539861) mouse Th2 cells [09-002]. (lymph) | SRR306542(GSM750585) 19-24nt. (ago2 brain) | SRR306534(GSM750577) 19-24nt. (ago2 brain) | SRR345204(SRX097265) source: size fractionated RNA from mouse hipp. (brain) | SRR346423(SRX098265) Global profiling of miRNA and the hairpin pre. (Brain) | SRR346424(SRX098266) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR042458(GSM539850) mouse basophil cells [09-002]. (b cell) | SRR059769(GSM562831) Treg_Drosha. (spleen) | SRR553602(SRX182808) source: Sertoli cells. (Sertoli cells) | SRR279907(GSM689057) cell type: mouse embryonic stem cellcell line. (ESC) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR042462(GSM539854) mouse macrophages [09-002]. (bone marrow) | SRR346415(SRX098256) source: Testis. (Testes) | SRR037905(GSM510441) brain_rep2. (brain) | SRR391847(GSM852142) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR037937(GSM510475) 293cand2. (cell line) | SRR039183(GSM485323) shLuc.1309 (-dox). (fibroblast) | SRR116846(GSM678422) Small RNA deep sequencing from mouse epididym. (epididymis) | SRR306536(GSM750579) 19-24nt. (ago2 brain) | SRR206941(GSM723282) other. (brain) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR685339(GSM1079783) Small RNAs (15-50 nts in length) from immorta. (cell line) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR042444(GSM539836) mouse pre B cells [09-002]. (b cell) | SRR306530(GSM750573) 19-24nt. (ago2 brain) | SRR073954(GSM629280) total RNA. (blood) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR346422(SRX098264) Global profiling of miRNA and the hairpin pre. (Muscle) | SRR206940(GSM723281) other. (brain) | SRR306540(GSM750583) 19-24nt. (ago2 brain) | GSM314557(GSM314557) ESC dgcr8 (Illumina). (ESC) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | GSM519104(GSM519104) mouse_granulocyte_nuclei_smallRNAs. (cell line) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM510444(GSM510444) brain_rep5. (brain) | mjTestesWT2() Testes Data. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR038741(GSM527276) small RNA-Seq. (brain) | SRR345205(SRX097266) source: size fractionated RNA from mouse hipp. (brain) | SRR039185(GSM485325) shLuc.1309;ROSA-rtTA (-dox). (fibroblast) | SRR346417(SRX098258) Global profiling of miRNA and the hairpin pre. (Brain) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (ago2 liver) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR042451(GSM539843) mouse B cells activated in vitro with LPS/aIgD-Dextran [09-002]. (b cell) | SRR029123(GSM416611) NIH3T3. (cell line) | SRR042443(GSM539835) mouse pro B cells [09-002]. (b cell) | SRR525239(SRA056111/SRX170315) Mus musculus domesticus miRNA sequencing. (ago1 Blood) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR042452(GSM539844) mouse B1 B cells [09-002]. (b cell) | SRR553586(SRX182792) source: Testis. (testes) | SRR553583(SRX182789) source: Cerebellum. (Cerebellum) | SRR346414(SRX098255) Global profiling of miRNA and the hairpin pre. (Spleen) | SRR059765(GSM562827) DN3_control. (thymus) | mjTestesWT3() Testes Data. (testes) | SRR059766(GSM562828) DN3_Drosha. (thymus) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR306544(GSM750587) 19-24nt. (ago2 brain) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................................................ACGTGACCATTTCTGCCCAATT.................................................... | 22 | 1 | 26.00 | 26.00 | 7.00 | - | 2.00 | - | - | - | 3.00 | - | - | - | - | 2.00 | - | 1.00 | 1.00 | 1.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................ACGTGACCATTTCTGCCCAAT..................................................... | 21 | 1 | 12.00 | 12.00 | 3.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | 1.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TGACGTGACCATTTCTGCCCAAT..................................................... | 23 | 1 | 9.00 | 9.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .................................................................................................................................CTGGGGAGAATGGCCATTTCA.................................................................................................... | 21 | 1 | 7.00 | 7.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - |
| .................................................................................................................................CTGGGGAGAATGGCCATTTCAA................................................................................................... | 22 | 1 | 5.00 | 7.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................GCATGGGAATATTGGCG...................................................................................................................................................................................................... | 17 | 5.00 | 0.00 | - | 2.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................CTGGGGAGAATGGCCATTTC..................................................................................................... | 20 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................TGGCGAAACGGAAGCGCAGTGACTCC........... | 26 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................CTGGGGAGAATGGCCATTTCAT................................................................................................... | 22 | 1 | 4.00 | 4.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................TCCTGGGGAGAATGGCCATTTC..................................................................................................... | 22 | 1 | 3.00 | 3.00 | - | - | - | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................ACGTGACCATTTCTGCCAATT..................................................... | 21 | 3.00 | 0.00 | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................TGACGTGACCATTTCTGCCCAA...................................................... | 22 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TGACGTGACCATTTCTGCCCAAC..................................................... | 23 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................CCTGGGGAGAATGGCCATTTCA.................................................................................................... | 22 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................CTGGGGAGAATGGCCATTT...................................................................................................... | 19 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TGACGTGACCATTTCTGCCCAATT.................................................... | 24 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................TCCTGGGGAGAATGGCCATT....................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................CTGGGGAGAATGGCCATT....................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................CCTGGGGAGAATGGCCATT....................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................ACGTGACCATTTCTGCCCACTC.................................................... | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................................ACGTGACCATTTCTGCCCCAT..................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................................................................GTCCCTGGCGAAACGGAAGCGC.................... | 22 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................GGGTGTGTACTTCAGTGTG.................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................................ACGTGACCATTTCTGCCCAAC..................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................GCATGGGAATATTGGCGC..................................................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................TGGGGAGAATGGCCATTTCA.................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................GGAAGCGCAGTGACTCCG.......... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................CTGACGTGACCATTTCTGCCCAAA..................................................... | 24 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................GCATGGGAATATTGGTG...................................................................................................................................................................................................... | 17 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................TCCTGGGGAGAATGGCCATTT...................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................CGGAAGCGCAGTGACTCCGAGGAGAAGG. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ..................................................................................................................................TGGGGAGAATGGCCATTTTA.................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................................................................................................TGGCGAAACGGAAGCGCAGTGACTTC........... | 26 | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................................TTTTCTGGTGAGCAATCCC........................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................................ACGTGACCATTTCTGCCCAATTC................................................... | 23 | 1 | 1.00 | 26.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................TGGGAATATTGGTGACATG................................................................................................................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................TGACGTGACCATTTCTGCCCGAC..................................................... | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................GGCGTGTAGAAAAAAAA............................................................................................................................ | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................................ACGTGACCATTTCTGCCCAATTT................................................... | 23 | 1 | 1.00 | 26.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................GCGTGTAGAAAAAAAA............................................................................................................................ | 16 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | |
| ......................................................................GCTCAGTGCTTATGGGTGTGTACTTCAGGTGA.................................................................................................................................................... | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................TGGGGAGAATGGCCATTTCAT................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................ACGTGACCATTTCTGCCCATT..................................................... | 21 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................GGCGTGTAGAAAAAAAAAA.......................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................TGACGTGACCATTTCTGCCCTTG..................................................... | 23 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................................................................................AAACGGAAGCGCAG.................. | 14 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - |
| ......................................................................................................................................................................................................AGGAGCTCTTGTCCCTG................................... | 17 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - |
| CCCTTAAAGGAGGACAGATTTTGGAGTTTAAGTTTGCATGGGAATATTGGTGATCTGTCTCTGCTCTTGTGCTCAGTGCTTATGGGTGTGTACTTCAGGTGAAAGCACAGGCGTGTAGAAAAAAAGATCCTGGGGAGAATGGCCATTTCATTACACTTTTCTGGTGAGCAAAACTGACGTGACCATTTCTGCCCAATTAGGAGCTCTTGTCCCTGGCGAAACGGAAGCGCAGTGACTCCGAGGAGAAGGA ..................................................................................................................................((((.(((((((.(((.(((...((((.....))))........))).))).))))).)).))))....................................................... ............................................................................................................................125........................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR546155(SRX180174) Global profiling of miRNA and the hairpin pre. (ago2 Neuroblastoma) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR038744(GSM527279) small RNA-Seq. (brain) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR306537(GSM750580) 19-24nt. (ago2 brain) | mjLiverKO1() Liver Data. (Zcchc11 liver) | SRR553582(SRX182788) source: Brain. (Brain) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | SRR042469(GSM539861) mouse Th2 cells [09-002]. (lymph) | SRR306542(GSM750585) 19-24nt. (ago2 brain) | SRR306534(GSM750577) 19-24nt. (ago2 brain) | SRR345204(SRX097265) source: size fractionated RNA from mouse hipp. (brain) | SRR346423(SRX098265) Global profiling of miRNA and the hairpin pre. (Brain) | SRR346424(SRX098266) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR042458(GSM539850) mouse basophil cells [09-002]. (b cell) | SRR059769(GSM562831) Treg_Drosha. (spleen) | SRR553602(SRX182808) source: Sertoli cells. (Sertoli cells) | SRR279907(GSM689057) cell type: mouse embryonic stem cellcell line. (ESC) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR042462(GSM539854) mouse macrophages [09-002]. (bone marrow) | SRR346415(SRX098256) source: Testis. (Testes) | SRR037905(GSM510441) brain_rep2. (brain) | SRR391847(GSM852142) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR037937(GSM510475) 293cand2. (cell line) | SRR039183(GSM485323) shLuc.1309 (-dox). (fibroblast) | SRR116846(GSM678422) Small RNA deep sequencing from mouse epididym. (epididymis) | SRR306536(GSM750579) 19-24nt. (ago2 brain) | SRR206941(GSM723282) other. (brain) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR685339(GSM1079783) Small RNAs (15-50 nts in length) from immorta. (cell line) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR037902(GSM510438) testes_rep3. (testes) | SRR042444(GSM539836) mouse pre B cells [09-002]. (b cell) | SRR306530(GSM750573) 19-24nt. (ago2 brain) | SRR073954(GSM629280) total RNA. (blood) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR346422(SRX098264) Global profiling of miRNA and the hairpin pre. (Muscle) | SRR206940(GSM723281) other. (brain) | SRR306540(GSM750583) 19-24nt. (ago2 brain) | GSM314557(GSM314557) ESC dgcr8 (Illumina). (ESC) | SRR069810(GSM610966) small RNA sequencing; sample 2. (testes) | GSM519104(GSM519104) mouse_granulocyte_nuclei_smallRNAs. (cell line) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM510444(GSM510444) brain_rep5. (brain) | mjTestesWT2() Testes Data. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR038741(GSM527276) small RNA-Seq. (brain) | SRR345205(SRX097266) source: size fractionated RNA from mouse hipp. (brain) | SRR039185(GSM485325) shLuc.1309;ROSA-rtTA (-dox). (fibroblast) | SRR346417(SRX098258) Global profiling of miRNA and the hairpin pre. (Brain) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (ago2 liver) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR042451(GSM539843) mouse B cells activated in vitro with LPS/aIgD-Dextran [09-002]. (b cell) | SRR029123(GSM416611) NIH3T3. (cell line) | SRR042443(GSM539835) mouse pro B cells [09-002]. (b cell) | SRR525239(SRA056111/SRX170315) Mus musculus domesticus miRNA sequencing. (ago1 Blood) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | GSM475280(GSM475280) Mili-IP. (mili testes) | SRR042452(GSM539844) mouse B1 B cells [09-002]. (b cell) | SRR553586(SRX182792) source: Testis. (testes) | SRR553583(SRX182789) source: Cerebellum. (Cerebellum) | SRR346414(SRX098255) Global profiling of miRNA and the hairpin pre. (Spleen) | SRR059765(GSM562827) DN3_control. (thymus) | mjTestesWT3() Testes Data. (testes) | SRR059766(GSM562828) DN3_Drosha. (thymus) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR306544(GSM750587) 19-24nt. (ago2 brain) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................................................................................TCTTGTCCCTGGCGAACAG........................... | 19 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................TTTCTGGTGAGCAAAA............................................................................. | 16 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 |