| (2) AGO1.ip | (4) AGO2.ip | (1) AGO3.ip | (21) BRAIN | (2) CELL-LINE | (1) DCR.mut | (2) DGCR8.mut | (10) EMBRYO | (3) ESC | (2) FIBROBLAST | (4) HEART | (2) KIDNEY | (7) LIVER | (2) LUNG | (6) OTHER | (6) OTHER.mut | (2) PIWI.ip | (3) SKIN | (13) TESTES | (3) TOTAL-RNA |
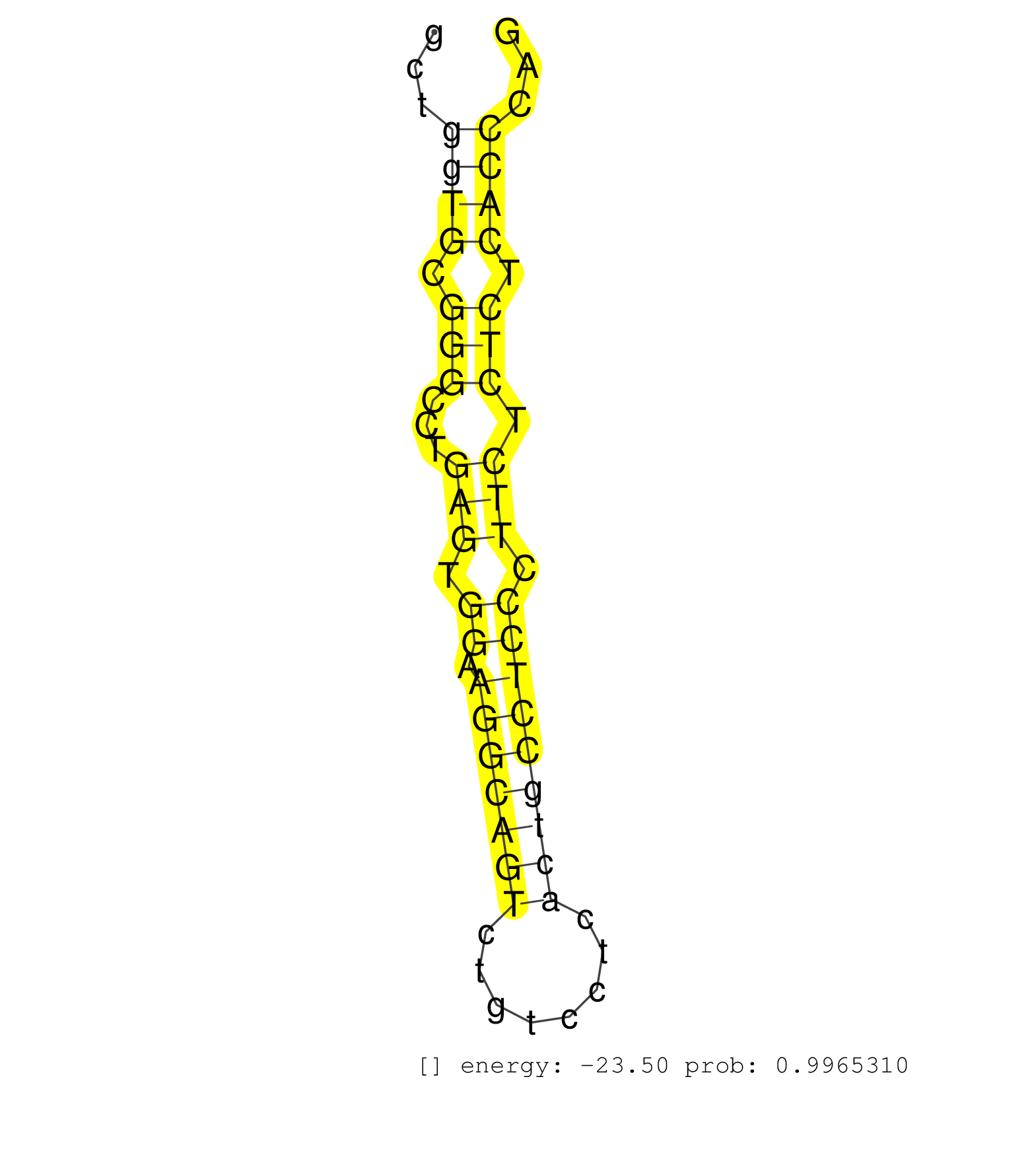
| TGCACTTCATCCAGATTCTTCTAGGTGTTTGTTTGTGTACCACTTCCCTTATTAGGCTGCTTCTGTCAGTTCCTGAGGGTAGGAAGTGAGTTCCAGGTCTGACTGCTTCAGGTCTAGCCTATGGCTATGCTCAGAGAATGCTGGTGCGGGCCTGAGTGGAAGGCAGTCTGTCCTCACTGCCTCCCTTCTCTCTCACCCAGATGAAAACCCAGTATGAGCAGACCCTGGCCGAGCTAAATCGCTACACCCC ..............................................................................................................................................((((.(((...(((.((.(((((((........))))))))).))).))).))))..................................................... ...........................................................................................................................................140.........................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR279907(GSM689057) cell type: mouse embryonic stem cellcell line. (ESC) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | Ago1IP517(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | mjLiverKO1() Liver Data. (Zcchc11 liver) | SRR345203(SRX097264) source: size fractionated RNA from mouse hipp. (brain) | SRR039186(GSM485326) shLuc.1309;ROSA-rtTA (+dox). (fibroblast) | SRR346417(SRX098258) Global profiling of miRNA and the hairpin pre. (Brain) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR346422(SRX098264) Global profiling of miRNA and the hairpin pre. (Muscle) | SRR553583(SRX182789) source: Cerebellum. (Cerebellum) | SRR038745(GSM527280) small RNA-Seq. (dgcr8 brain) | mjLiverWT1() Liver Data. (liver) | SRR345202(SRX097263) source: size fractionated RNA from mouse hipp. (brain) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjLiverKO2() Liver Data. (Zcchc11 liver) | mjTestesWT4() Testes Data. (testes) | SRR553584(SRX182790) source: Heart. (Heart) | mjTestesWT3() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR345205(SRX097266) source: size fractionated RNA from mouse hipp. (brain) | SRR042474(GSM539866) mouse embryonic stem cells [09-002]. (ESC) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) | SRR553585(SRX182791) source: Kidney. (Kidney) | SRR553602(SRX182808) source: Sertoli cells. (Sertoli cells) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT2() Testes Data. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR038743(GSM527278) small RNA-Seq. (dicer brain) | SRR345207(SRX097268) source: size fractionated RNA from mouse hipp. (brain) | SRR039610(GSM527274) small RNA-Seq. (brain) | SRR346419(SRX098260) Global profiling of miRNA and the hairpin pre. (Lung) | SRR037901(GSM510437) testes_rep2. (testes) | SRR042478(GSM539870) mouse lung tissue [09-002]. (lung) | mjLiverWT2() Liver Data. (liver) | GSM510452(GSM510452) newborn_rep8. (total RNA) | SRR059776(GSM562838) MEF_Dicer. (MEF) | SRR553586(SRX182792) source: Testis. (testes) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | Ago1IP812(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | GSM640582(GSM640582) small RNA in the liver with paternal control . (liver) | GSM640578(GSM640578) small RNA in the liver with paternal control. (liver) | mjLiverKO3() Liver Data. (Zcchc11 liver) | SRR038741(GSM527276) small RNA-Seq. (brain) | SRR553582(SRX182788) source: Brain. (Brain) | SRR206939(GSM723280) other. (brain) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR037920(GSM510458) e12p5_rep2. (embryo) | SRR042476(GSM539868) mouse heart tissue [09-002]. (heart) | SRR042484(GSM539876) mouse salivary gland tissue [09-002]. (salivary gland) | SRR306532(GSM750575) 19-24nt. (ago2 brain) | SRR095855BC1(SRX039178) sequencing of miRNA from wild type and diseas. (heart) | GSM510450(GSM510450) newborn_rep6. (total RNA) | SRR037937(GSM510475) 293cand2. (cell line) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR042475(GSM539867) mouse embryonic fibroblast cells [09-002]. (fibroblast) | SRR306540(GSM750583) 19-24nt. (ago2 brain) | SRR042483(GSM539875) mouse muscle tissue [09-002]. (muscle) | GSM519104(GSM519104) mouse_granulocyte_nuclei_smallRNAs. (cell line) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR306536(GSM750579) 19-24nt. (ago2 brain) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | GSM475281(GSM475281) total RNA. (testes) | SRR345204(SRX097265) source: size fractionated RNA from mouse hipp. (brain) | GSM510456(GSM510456) newborn_rep12. (total RNA) | SRR346423(SRX098265) Global profiling of miRNA and the hairpin pre. (Brain) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | Ago3IP805(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | SRR346416(SRX098257) Global profiling of miRNA and the hairpin pre. (Kidney) | SRR206940(GSM723281) other. (brain) | SRR095855BC7(SRX039178) sequencing of miRNA from wild type and diseas. (heart) | SRR037924(GSM510462) e9p5_rep2. (embryo) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................TGCGGGCCTGAGTGGAAGGCAGT................................................................................... | 23 | 1 | 29.00 | 29.00 | - | 1.00 | - | - | - | 1.00 | 2.00 | - | - | - | 2.00 | - | 2.00 | 2.00 | - | - | - | - | 1.00 | 1.00 | - | - | 1.00 | - | - | 1.00 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | 1.00 | - | - | 1.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | 1.00 | 1.00 | 1.00 | - | - | - | - | - |
| ................................................................................................................................................TGCGGGCCTGAGTGGAAGGCAG.................................................................................... | 22 | 1 | 18.00 | 18.00 | 4.00 | - | - | - | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 2.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...................................................................................................................................................................................CCTCCCTTCTCTCTCACCCAGT................................................. | 22 | 10.00 | 0.00 | - | - | 5.00 | - | - | - | 1.00 | - | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................TGCGGGCCTGAGTGGAAGGCAGA................................................................................... | 23 | 1 | 9.00 | 18.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | 2.00 | - | 1.00 | - | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................CTGGTGCGGGCCTGAGTGGAAGGCA..................................................................................... | 25 | 1 | 5.00 | 5.00 | - | - | - | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................CGGGCCTGAGTGGAAGGCAGT................................................................................... | 21 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TGCCTCCCTTCTCTCTCACCCAGT................................................. | 24 | 4.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | |
| .................................................................................................................................................GCGGGCCTGAGTGGAAGGCAGT................................................................................... | 22 | 1 | 3.00 | 3.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................TGCGGGCCTGAGTGGAAGGCAT.................................................................................... | 22 | 1 | 3.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TGGTGCGGGCCTGAGTGGAAGGC...................................................................................... | 23 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................TGCGGGCCTGAGTGGAAGGCA..................................................................................... | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................TGCGGGCCTGAGTGGAAGGCAGTT.................................................................................. | 24 | 1 | 2.00 | 29.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................CGGGCCTGAGTGGAAGGCAGTAA................................................................................. | 23 | 1 | 2.00 | 4.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TGGTGCGGGCCTGAGTGGAAGGCA..................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................TGCGGGCCTGAGTGGAAGG....................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................TGCGGGCCTGAGTGGAAGGCAGG................................................................................... | 23 | 1 | 1.00 | 18.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................GGCCTGAGTGGAAGGGA..................................................................................... | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................TGCGGGCCTGAGTGGAAGGCAA.................................................................................... | 22 | 1 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................TGCGGGCCTGAGTGGAAG........................................................................................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TGGTGCGGGCCTGAGTGGAAGGCAGT................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................GCTGGTGCGGGCCTGTG.............................................................................................. | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................TGCGGGCCTGAGTGGAAGGCAGTAT................................................................................. | 25 | 1 | 1.00 | 29.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TGGTGCGGGCCTGAGTGGAAGG....................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................GCGGGCCTGAGTGGAAGGA...................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...................................................................................................................................................................................CCTCCCTTCTCTCTCACCCAGA................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .................................................................................................................................................................................TGCCTCCCTTCTCTCTCACCCAGA................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TGGTGCGGGCCTGAGTGGAAGGCAG.................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................................TATGAGCAGACCCTGGCCGAGCTAAATCGC........ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................GTGCGGGCCTGAGTGGAAGGCAGT................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................GTGCGGGCCTGAGTGGAAGGCA..................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................................AGCAGACCCTGGCCGAGCTAAATCGC........ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .................................................................................................................................................GCGGGCCTGAGTGGAAGG....................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................GCCTCCCTTCTCTCTCACCCAGT................................................. | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................AGTGGAAGGCAGTCTGTTT............................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................GCGGGCCTGAGTGGAAGGCAG.................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................CTCCCTTCTCTCTCACCCAGA................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TGGTGCGGGCCTGAGTGGAA......................................................................................... | 20 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................TTCCTGAGGGTAGGAGGA................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................GGTGCGGGCCTGAGTGGAAG........................................................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................TGCGGGCCTGAGTGGAAGGC...................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................AGAGAATGCTGGTGCG...................................................................................................... | 16 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TGCACTTCATCCAGATTCTTCTAGGTGTTTGTTTGTGTACCACTTCCCTTATTAGGCTGCTTCTGTCAGTTCCTGAGGGTAGGAAGTGAGTTCCAGGTCTGACTGCTTCAGGTCTAGCCTATGGCTATGCTCAGAGAATGCTGGTGCGGGCCTGAGTGGAAGGCAGTCTGTCCTCACTGCCTCCCTTCTCTCTCACCCAGATGAAAACCCAGTATGAGCAGACCCTGGCCGAGCTAAATCGCTACACCCC ..............................................................................................................................................((((.(((...(((.((.(((((((........))))))))).))).))).))))..................................................... ...........................................................................................................................................140.........................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR279907(GSM689057) cell type: mouse embryonic stem cellcell line. (ESC) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | Ago1IP517(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | mjLiverKO1() Liver Data. (Zcchc11 liver) | SRR345203(SRX097264) source: size fractionated RNA from mouse hipp. (brain) | SRR039186(GSM485326) shLuc.1309;ROSA-rtTA (+dox). (fibroblast) | SRR346417(SRX098258) Global profiling of miRNA and the hairpin pre. (Brain) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR346422(SRX098264) Global profiling of miRNA and the hairpin pre. (Muscle) | SRR553583(SRX182789) source: Cerebellum. (Cerebellum) | SRR038745(GSM527280) small RNA-Seq. (dgcr8 brain) | mjLiverWT1() Liver Data. (liver) | SRR345202(SRX097263) source: size fractionated RNA from mouse hipp. (brain) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | mjLiverKO2() Liver Data. (Zcchc11 liver) | mjTestesWT4() Testes Data. (testes) | SRR553584(SRX182790) source: Heart. (Heart) | mjTestesWT3() Testes Data. (testes) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | SRR345205(SRX097266) source: size fractionated RNA from mouse hipp. (brain) | SRR042474(GSM539866) mouse embryonic stem cells [09-002]. (ESC) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) | SRR553585(SRX182791) source: Kidney. (Kidney) | SRR553602(SRX182808) source: Sertoli cells. (Sertoli cells) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | mjTestesWT2() Testes Data. (testes) | mjTestesKO5() Testes Data. (Zcchc11 testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR038743(GSM527278) small RNA-Seq. (dicer brain) | SRR345207(SRX097268) source: size fractionated RNA from mouse hipp. (brain) | SRR039610(GSM527274) small RNA-Seq. (brain) | SRR346419(SRX098260) Global profiling of miRNA and the hairpin pre. (Lung) | SRR037901(GSM510437) testes_rep2. (testes) | SRR042478(GSM539870) mouse lung tissue [09-002]. (lung) | mjLiverWT2() Liver Data. (liver) | GSM510452(GSM510452) newborn_rep8. (total RNA) | SRR059776(GSM562838) MEF_Dicer. (MEF) | SRR553586(SRX182792) source: Testis. (testes) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | Ago1IP812(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | GSM640582(GSM640582) small RNA in the liver with paternal control . (liver) | GSM640578(GSM640578) small RNA in the liver with paternal control. (liver) | mjLiverKO3() Liver Data. (Zcchc11 liver) | SRR038741(GSM527276) small RNA-Seq. (brain) | SRR553582(SRX182788) source: Brain. (Brain) | SRR206939(GSM723280) other. (brain) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR037920(GSM510458) e12p5_rep2. (embryo) | SRR042476(GSM539868) mouse heart tissue [09-002]. (heart) | SRR042484(GSM539876) mouse salivary gland tissue [09-002]. (salivary gland) | SRR306532(GSM750575) 19-24nt. (ago2 brain) | SRR095855BC1(SRX039178) sequencing of miRNA from wild type and diseas. (heart) | GSM510450(GSM510450) newborn_rep6. (total RNA) | SRR037937(GSM510475) 293cand2. (cell line) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR042475(GSM539867) mouse embryonic fibroblast cells [09-002]. (fibroblast) | SRR306540(GSM750583) 19-24nt. (ago2 brain) | SRR042483(GSM539875) mouse muscle tissue [09-002]. (muscle) | GSM519104(GSM519104) mouse_granulocyte_nuclei_smallRNAs. (cell line) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR306536(GSM750579) 19-24nt. (ago2 brain) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | GSM475281(GSM475281) total RNA. (testes) | SRR345204(SRX097265) source: size fractionated RNA from mouse hipp. (brain) | GSM510456(GSM510456) newborn_rep12. (total RNA) | SRR346423(SRX098265) Global profiling of miRNA and the hairpin pre. (Brain) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | Ago3IP805(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | SRR346416(SRX098257) Global profiling of miRNA and the hairpin pre. (Kidney) | SRR206940(GSM723281) other. (brain) | SRR095855BC7(SRX039178) sequencing of miRNA from wild type and diseas. (heart) | SRR037924(GSM510462) e9p5_rep2. (embryo) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................AGGCTGCTTCTGTCAGTTCCTGAGGGTA......................................................................................................................................................................... | 28 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................GTCTAGCCTATGGCTATGCTCAGAGAA................................................................................................................ | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................CCCTTCTCTCTCACCCCATT................................................ | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .GCACTTCATCCAGATTCTTCTA................................................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................GGCTGCTTCTGTCAGTTCCTGAGGGTA......................................................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TGCCTCCCTTCTCTCTCACC..................................................... | 20 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................GTGAGTTCCAGGTCTGGTC.................................................................................................................................................. | 19 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .GCACTTCATCCAGATTCTTCT.................................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................CCCTTCTCTCTCACCCTGA................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................CCAGGTCTGACTGCTTCAGGTCTAGCCTA................................................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................................................GAAGTGAGTTCCAGGTCTT..................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................TTGTTTGTGTACCACTTCCCTTATTA.................................................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................CTTCTCTCTCACCCACAGA............................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................................AAGGCAGTCTGTCCTC........................................................................... | 16 | 9 | 0.11 | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |