| (1) AGO1.ip | (12) AGO2.ip | (4) B-CELL | (28) BRAIN | (1) CELL-LINE | (2) DGCR8.mut | (3) EMBRYO | (3) ESC | (1) KIDNEY | (2) LIVER | (1) LUNG | (4) LYMPH | (10) OTHER | (1) OTHER.mut | (1) PANCREAS | (7) SPLEEN | (3) TESTES | (1) THYMUS |
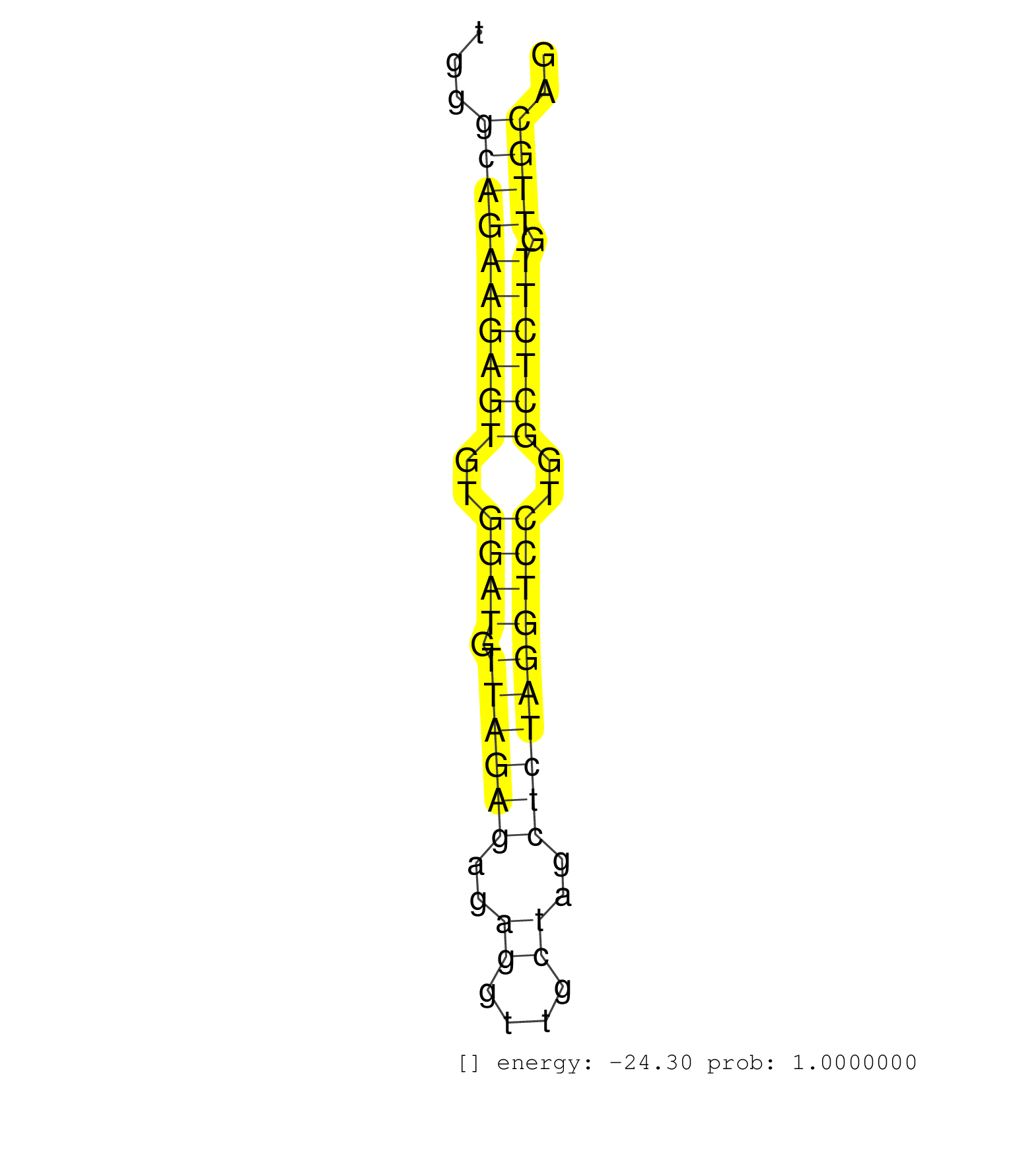
| GCGCAGTGCAGTCAGAGGATGACGCCCGGCAGGATGTCATCCAAGACGTGGTAAGCGTCTGGGCAAGGGATTGGGATCAGCTGGGCAGAAGAGTGTGGATGTTAGAGAGAGGTTGCTAGCTCTAGGTCCTGGCTCTTGTTGCAGGAGATCAGCCGGAAGGTGTACAAGGGGATGCTAGACCTCCTCAAGTGCAC ....................................................................................((((((((((..((((.((((((..((....))..))))))))))..)))))).)))).................................................... .................................................................................82............................................................144................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR546155(SRX180174) Global profiling of miRNA and the hairpin pre. (ago2 Neuroblastoma) | SRR346423(SRX098265) Global profiling of miRNA and the hairpin pre. (Brain) | SRR306542(GSM750585) 19-24nt. (ago2 brain) | SRR553582(SRX182788) source: Brain. (Brain) | SRR306544(GSM750587) 19-24nt. (ago2 brain) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR346421(SRX098263) Global profiling of miRNA and the hairpin pre. (mixture) | SRR553583(SRX182789) source: Cerebellum. (Cerebellum) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | SRR345201(SRX097262) source: size fractionated RNA from mouse hipp. (brain) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) | SRR346417(SRX098258) Global profiling of miRNA and the hairpin pre. (Brain) | SRR206942(GSM723283) other. (brain) | mjLiverKO3() Liver Data. (Zcchc11 liver) | SRR039610(GSM527274) small RNA-Seq. (brain) | SRR042466(GSM539858) mouse CD8+ T cells (spleen) [09-002]. (spleen) | SRR042471(GSM539863) mouse TFHS cells [09-002]. (spleen) | SRR038744(GSM527279) small RNA-Seq. (brain) | SRR346414(SRX098255) Global profiling of miRNA and the hairpin pre. (Spleen) | SRR306534(GSM750577) 19-24nt. (ago2 brain) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR306532(GSM750575) 19-24nt. (ago2 brain) | SRR346425(SRX098267) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR345205(SRX097266) source: size fractionated RNA from mouse hipp. (brain) | SRR038741(GSM527276) small RNA-Seq. (brain) | SRR042465(GSM539857) mouse CD4+ T cells (spleen) [09-002]. (spleen) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR042454(GSM539846) mouse germinal center B cells (lymph node) [09-002]. (b cell) | SRR206940(GSM723281) other. (brain) | SRR306536(GSM750579) 19-24nt. (ago2 brain) | mjLiverWT1() Liver Data. (liver) | SRR042469(GSM539861) mouse Th2 cells [09-002]. (lymph) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR306533(GSM750576) 19-24nt. (ago2 brain) | SRR346419(SRX098260) Global profiling of miRNA and the hairpin pre. (Lung) | SRR042467(GSM539859) mouse T cells activated in vitro with Concanavalin A [09-002]. (spleen) | SRR279906(GSM689056) cell type: mouse embryonic stem cellcell line. (ESC) | SRR059772(GSM562834) CD4_Drosha. (spleen) | SRR039152(GSM471929) HL1_siNon_smallRNAseq. (muscle) | SRR525239(SRA056111/SRX170315) Mus musculus domesticus miRNA sequencing. (ago1 Blood) | SRR037901(GSM510437) testes_rep2. (testes) | SRR042474(GSM539866) mouse embryonic stem cells [09-002]. (ESC) | SRR037905(GSM510441) brain_rep2. (brain) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR042464(GSM539856) mouse CD4+CD8+ T cells (thymus) [09-002]. (thymus) | SRR206939(GSM723280) other. (brain) | SRR038745(GSM527280) small RNA-Seq. (dgcr8 brain) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM510444(GSM510444) brain_rep5. (brain) | GSM519104(GSM519104) mouse_granulocyte_nuclei_smallRNAs. (cell line) | SRR306531(GSM750574) 19-24nt. (ago2 brain) | SRR042472(GSM539864) mouse iTreg cells [09-002]. (lymph) | SRR059774(GSM562836) MEF_control. (MEF) | SRR306540(GSM750583) 19-24nt. (ago2 brain) | SRR042446(GSM539838) mouse mature B cells (spleen) replicate 1 [09-002]. (b cell) | SRR042468(GSM539860) mouse Th1 cells [09-002]. (lymph) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR059769(GSM562831) Treg_Drosha. (spleen) | SRR039153(GSM471930) HL1_siSrf1_smallRNAseq. (muscle) | SRR306543(GSM750586) 19-24nt. (ago2 brain) | SRR306527(GSM750570) 19-24nt. (ago2 brain) | SRR042447(GSM539839) mouse mature B cells (spleen) replicate 2 [09-002]. (b cell) | SRR042457(GSM539849) mouse mast cells [09-002]. (bone marrow) | SRR346416(SRX098257) Global profiling of miRNA and the hairpin pre. (Kidney) | SRR279904(GSM689054) cell type: mouse embryonic stem cellcell line. (ESC) | SRR042458(GSM539850) mouse basophil cells [09-002]. (b cell) | SRR346424(SRX098266) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR065059(SRR065059) Tissue-specific Regulation of Mouse MicroRNA . (pancreas) | SRR042470(GSM539862) mouse Th17 cells [09-002]. (lymph) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................TAGGTCCTGGCTCTTGTTGCAGT................................................. | 23 | 1 | 65.00 | 38.00 | 22.00 | 6.00 | - | 6.00 | - | 5.00 | - | 1.00 | - | - | - | - | 2.00 | 3.00 | 2.00 | 1.00 | - | 1.00 | 1.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | 1.00 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | 1.00 | - | 1.00 | 1.00 | - |
| ..........................................................................................................................TAGGTCCTGGCTCTTGTTGCAG.................................................. | 22 | 1 | 38.00 | 38.00 | 2.00 | 8.00 | - | 3.00 | - | 4.00 | - | 5.00 | - | - | - | 4.00 | 2.00 | - | - | - | 1.00 | 1.00 | 1.00 | - | - | - | - | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ....AGTGCAGTCAGAGGAGCCA........................................................................................................................................................................... | 19 | 37.00 | 0.00 | - | - | 12.00 | - | 10.00 | - | - | - | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | |
| ......................................................................................AGAAGAGTGTGGATGTTAGA........................................................................................ | 20 | 1 | 10.00 | 10.00 | - | - | - | - | - | - | - | - | - | 6.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................TAGGTCCTGGCTCTTGTTGCAGA................................................. | 23 | 1 | 5.00 | 38.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..........................................................................................................................TAGGTCCTGGCTCTTGTTGC.................................................... | 20 | 1 | 5.00 | 5.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGCGTCTGGGCAAGGGATTGGGATCAGCTGGGCAGAAGAGTGTGGATGTTAGAGAGAGGTTGCTAGCTCTAGGTCCTGGCTCTTGTTGCAG.................................................. | 94 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................GGCAGAAGAGTGTGGATGTTA.......................................................................................... | 21 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................GGCAGAAGAGTGTGGATGTTAG......................................................................................... | 22 | 1 | 3.00 | 3.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....GTGCAGTCAGAGGATCCA........................................................................................................................................................................... | 18 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................................CCGGAAGGTGTACAAGGGGATGT................... | 23 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................GGAGATCAGCCGGAAGGTGT............................... | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGCGTCTGGGCAAGGGATTGGGATCAGCTGGGCAGAAGAGTGTGGATGTTAGAGAGAGGTTGCTAGCTCTAGGTCCTGGCTCTTGTTGCA................................................... | 93 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................AGGTCCTGGCTCTTGTTGCAGT................................................. | 22 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................TAGCTCTAGGTCCTGGCTCTTGT....................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| ..........................................................................................................................TAGGTCCTGGCTCTTGTTGCAT.................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................GCAGAAGAGTGTGGATGTTAG......................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....AGTGCAGTCAGAGGAGACA........................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................TAGAGAGAGGTTGCTAGCTCTAGGTCCTGG.............................................................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................CAGAAGAGTGTGGATGTTAGA........................................................................................ | 21 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAAGCGTCTGGGCAAGGGATTGGGATCAGCTGGGCAGAAGAGTGTGGATGTTAGAGAGAGGTTGCTAGCTCTAGGTCCTGGCTCTTGTTG..................................................... | 91 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................GGCAGAAGAGTGTGGATGTTAGA........................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................AGAAGAGTGTGGATGTTGTA........................................................................................ | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................TAGGTCCTGGCTCTTGTTGCA................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...CAGTGCAGTCAGAGGAGCCA........................................................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | |
| ...............................................GTGGTAAGCGTCTGGAGCT................................................................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................TAGGTCCTGGCTCTTGTTGCAGG................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................AAGCGTCTGGGCAAGGGATTGGGATCAGCTGGGCAGAAGAGTGTGGATGTTAGAGAGAGGTTGCTAGCTCTAGGTCCTGGCTCTTGTTGCAG.................................................. | 92 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................TAGGTCCTGGCTCTTGTTGCGGT................................................. | 23 | 1 | 1.00 | 5.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................TTGTTGCAGGAGATCAGTCGG...................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................GTCCTGGCTCTTGTTGCAG.................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................CTAGGTCCTGGCTCTTGTTGA.................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................TAGGTCCTGGCTCTTGTTGCAGTTT............................................... | 25 | 1 | 1.00 | 38.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ......................................................................................AGAAGAGTGTGGATGTTTGT........................................................................................ | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................CTTGTTGCAGGAGATGCC.......................................... | 18 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................GTAAGCGTCTGGGCAAGGGATTGGGATCAGCTGGGCAGAAGAGTGTGGATGTTAGAGAGAGGTTGCTAGCTCTAGGTCCTGGCTCTT......................................................... | 87 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................AGAAGAGTGTGGATGTTTGA........................................................................................ | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................GGAGATCAGCCGGAAGGT................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................TAGGTCCTGGCTCTTGTTGT.................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................TAGGTCCTGGCTCTTGTTGCAGTT................................................ | 24 | 1 | 1.00 | 38.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................GAAGGTGTACAAGGGGATG.................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................AGAAGAGTGTGGATGTTAAG........................................................................................ | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................CAGAAGAGTGTGGATGTTAG......................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................TAGGTCCTGGCTCTTGTTGCAAATA............................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................TAGGTCCTGGCTCTTGTTGCAA.................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................CAGAAGAGTGTGGATGTTTGAG....................................................................................... | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................CAGAAGAGTGTGGATGTTAGAGAGAGGTT................................................................................ | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................TGGATGTTAGAGAGAGTCT................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................AGAAGAGTGTGGATGTTAGT........................................................................................ | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................TAGGTCCTGGCTCTTGTTGCAGTAGA.............................................. | 26 | 1 | 1.00 | 38.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GCGCAGTGCAGTCAGAGGATGACGCCCGGCAGGATGTCATCCAAGACGTGGTAAGCGTCTGGGCAAGGGATTGGGATCAGCTGGGCAGAAGAGTGTGGATGTTAGAGAGAGGTTGCTAGCTCTAGGTCCTGGCTCTTGTTGCAGGAGATCAGCCGGAAGGTGTACAAGGGGATGCTAGACCTCCTCAAGTGCAC ....................................................................................((((((((((..((((.((((((..((....))..))))))))))..)))))).)))).................................................... .................................................................................82............................................................144................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR546155(SRX180174) Global profiling of miRNA and the hairpin pre. (ago2 Neuroblastoma) | SRR346423(SRX098265) Global profiling of miRNA and the hairpin pre. (Brain) | SRR306542(GSM750585) 19-24nt. (ago2 brain) | SRR553582(SRX182788) source: Brain. (Brain) | SRR306544(GSM750587) 19-24nt. (ago2 brain) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR346421(SRX098263) Global profiling of miRNA and the hairpin pre. (mixture) | SRR553583(SRX182789) source: Cerebellum. (Cerebellum) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | SRR345201(SRX097262) source: size fractionated RNA from mouse hipp. (brain) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) | SRR346417(SRX098258) Global profiling of miRNA and the hairpin pre. (Brain) | SRR206942(GSM723283) other. (brain) | mjLiverKO3() Liver Data. (Zcchc11 liver) | SRR039610(GSM527274) small RNA-Seq. (brain) | SRR042466(GSM539858) mouse CD8+ T cells (spleen) [09-002]. (spleen) | SRR042471(GSM539863) mouse TFHS cells [09-002]. (spleen) | SRR038744(GSM527279) small RNA-Seq. (brain) | SRR346414(SRX098255) Global profiling of miRNA and the hairpin pre. (Spleen) | SRR306534(GSM750577) 19-24nt. (ago2 brain) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR306532(GSM750575) 19-24nt. (ago2 brain) | SRR346425(SRX098267) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR345205(SRX097266) source: size fractionated RNA from mouse hipp. (brain) | SRR038741(GSM527276) small RNA-Seq. (brain) | SRR042465(GSM539857) mouse CD4+ T cells (spleen) [09-002]. (spleen) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR042454(GSM539846) mouse germinal center B cells (lymph node) [09-002]. (b cell) | SRR206940(GSM723281) other. (brain) | SRR306536(GSM750579) 19-24nt. (ago2 brain) | mjLiverWT1() Liver Data. (liver) | SRR042469(GSM539861) mouse Th2 cells [09-002]. (lymph) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR306533(GSM750576) 19-24nt. (ago2 brain) | SRR346419(SRX098260) Global profiling of miRNA and the hairpin pre. (Lung) | SRR042467(GSM539859) mouse T cells activated in vitro with Concanavalin A [09-002]. (spleen) | SRR279906(GSM689056) cell type: mouse embryonic stem cellcell line. (ESC) | SRR059772(GSM562834) CD4_Drosha. (spleen) | SRR039152(GSM471929) HL1_siNon_smallRNAseq. (muscle) | SRR525239(SRA056111/SRX170315) Mus musculus domesticus miRNA sequencing. (ago1 Blood) | SRR037901(GSM510437) testes_rep2. (testes) | SRR042474(GSM539866) mouse embryonic stem cells [09-002]. (ESC) | SRR037905(GSM510441) brain_rep2. (brain) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR042464(GSM539856) mouse CD4+CD8+ T cells (thymus) [09-002]. (thymus) | SRR206939(GSM723280) other. (brain) | SRR038745(GSM527280) small RNA-Seq. (dgcr8 brain) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM510444(GSM510444) brain_rep5. (brain) | GSM519104(GSM519104) mouse_granulocyte_nuclei_smallRNAs. (cell line) | SRR306531(GSM750574) 19-24nt. (ago2 brain) | SRR042472(GSM539864) mouse iTreg cells [09-002]. (lymph) | SRR059774(GSM562836) MEF_control. (MEF) | SRR306540(GSM750583) 19-24nt. (ago2 brain) | SRR042446(GSM539838) mouse mature B cells (spleen) replicate 1 [09-002]. (b cell) | SRR042468(GSM539860) mouse Th1 cells [09-002]. (lymph) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR059769(GSM562831) Treg_Drosha. (spleen) | SRR039153(GSM471930) HL1_siSrf1_smallRNAseq. (muscle) | SRR306543(GSM750586) 19-24nt. (ago2 brain) | SRR306527(GSM750570) 19-24nt. (ago2 brain) | SRR042447(GSM539839) mouse mature B cells (spleen) replicate 2 [09-002]. (b cell) | SRR042457(GSM539849) mouse mast cells [09-002]. (bone marrow) | SRR346416(SRX098257) Global profiling of miRNA and the hairpin pre. (Kidney) | SRR279904(GSM689054) cell type: mouse embryonic stem cellcell line. (ESC) | SRR042458(GSM539850) mouse basophil cells [09-002]. (b cell) | SRR346424(SRX098266) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR065059(SRR065059) Tissue-specific Regulation of Mouse MicroRNA . (pancreas) | SRR042470(GSM539862) mouse Th17 cells [09-002]. (lymph) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................TTGCAGGAGATCAGCAA....................................... | 17 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |