| (1) AGO.mut | (2) AGO1.ip | (4) AGO2.ip | (1) B-CELL | (7) BRAIN | (7) EMBRYO | (4) ESC | (1) HEART | (2) KIDNEY | (7) LIVER | (2) LUNG | (4) LYMPH | (5) OTHER | (6) OTHER.mut | (2) PIWI.ip | (1) PIWI.mut | (2) SKIN | (3) SPLEEN | (13) TESTES | (1) TOTAL-RNA | (3) UTERUS |
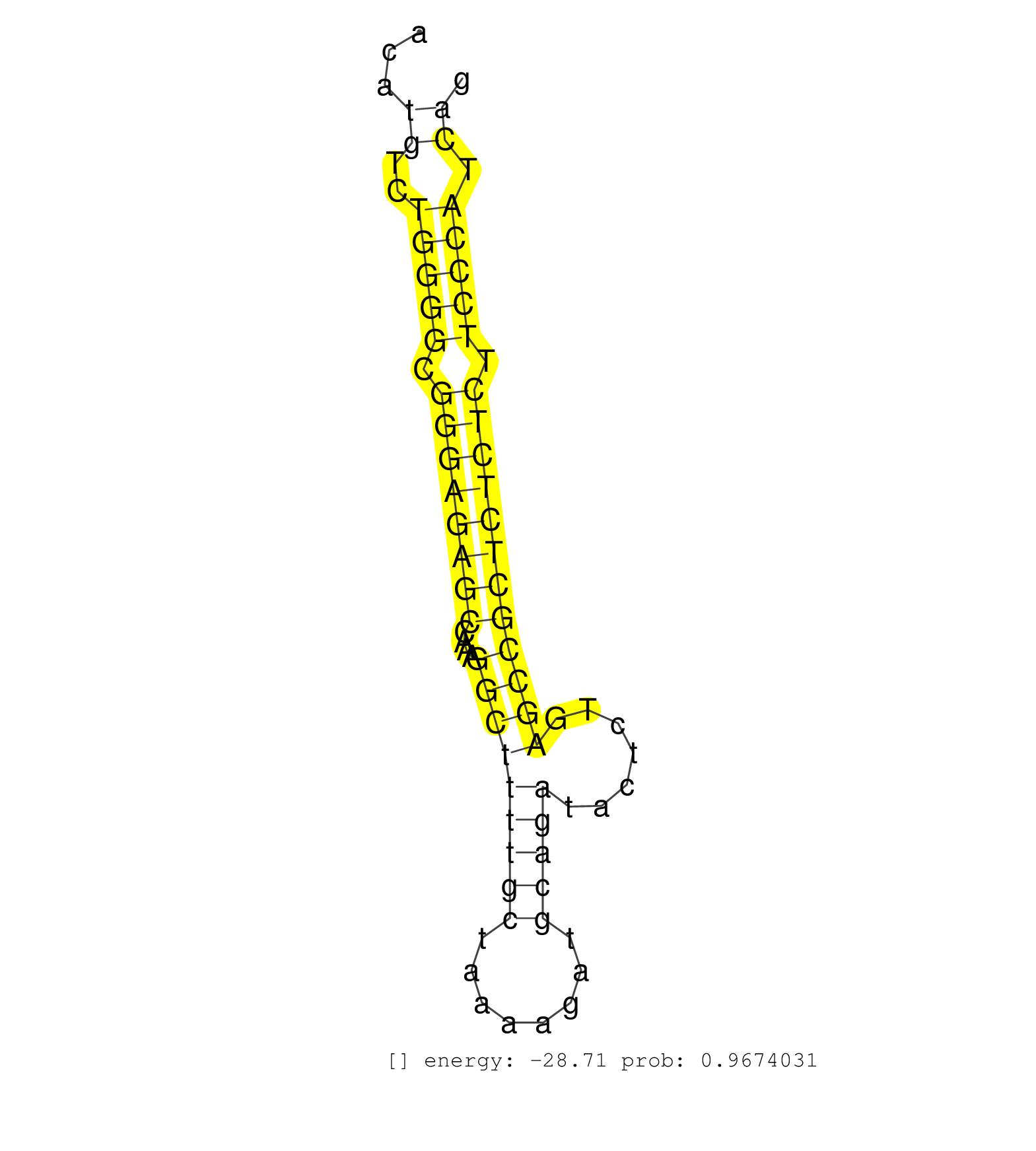
| GGAGCGAGCGAACAGCCCACAGGTTGAGGCTAACCTCACCGTCTCTGCTTCTGTGGCTCACAGAATTGGATTGTTTGTGTTTCTGCCTAACACTTCTCTGAGAGGTTTATCTACCAGGCTTCTGACATGTCTGGGGCGGGAGAGCCAAAGGCTTTTGCTAAAAGATGCAGATACTCTGAGCCGCTCTCTCTTCCCATCAGTGAGATCGAGAAGAACGACGTGGTGTTCTCCATGGACCAGACCGAATCCC ...............................................................................................................................((..(((((.((((((((....(((((((((........))))).......)))))))))))).))))).))................................................... ............................................................................................................................125........................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR346421(SRX098263) Global profiling of miRNA and the hairpin pre. (mixture) | mjTestesWT1() Testes Data. (testes) | mjLiverWT1() Liver Data. (liver) | mjLiverKO2() Liver Data. (Zcchc11 liver) | mjLiverKO1() Liver Data. (Zcchc11 liver) | GSM314557(GSM314557) ESC dgcr8 (Illumina). (ESC) | SRR346419(SRX098260) Global profiling of miRNA and the hairpin pre. (Lung) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR059769(GSM562831) Treg_Drosha. (spleen) | SRR346422(SRX098264) Global profiling of miRNA and the hairpin pre. (Muscle) | SRR032476(GSM485234) sRNA_delayed_deep_sequencing. (uterus) | mjLiverWT3() Liver Data. (liver) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR042463(GSM539855) mouse natural killer cells [09-002]. (spleen) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (ago2 liver) | SRR206942(GSM723283) other. (brain) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR346415(SRX098256) source: Testis. (Testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR042469(GSM539861) mouse Th2 cells [09-002]. (lymph) | mjLiverKO3() Liver Data. (Zcchc11 liver) | SRR346414(SRX098255) Global profiling of miRNA and the hairpin pre. (Spleen) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR553583(SRX182789) source: Cerebellum. (Cerebellum) | Ago1IP812(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR279903(GSM689053) cell type: mouse embryonic stem cellcell line. (ESC) | SRR346420(SRX098261) Global profiling of miRNA and the hairpin pre. (Uterus) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR306533(GSM750576) 19-24nt. (ago2 brain) | SRR032477(GSM485235) sRNA_activation_deep_sequencing. (uterus) | SRR042472(GSM539864) mouse iTreg cells [09-002]. (lymph) | SRR037926(GSM510464) e9p5_rep4. (embryo) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | Ago1IP517(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | SRR553585(SRX182791) source: Kidney. (Kidney) | SRR306540(GSM750583) 19-24nt. (ago2 brain) | GSM509276(GSM509276) small RNA cloning by length. (testes) | SRR037928(GSM510466) e7p5_rep2. (embryo) | SRR042468(GSM539860) mouse Th1 cells [09-002]. (lymph) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR042478(GSM539870) mouse lung tissue [09-002]. (lung) | GSM510454(GSM510454) newborn_rep10. (total RNA) | SRR042474(GSM539866) mouse embryonic stem cells [09-002]. (ESC) | SRR073954(GSM629280) total RNA. (blood) | SRR346416(SRX098257) Global profiling of miRNA and the hairpin pre. (Kidney) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR095855BC3(SRX039178) sequencing of miRNA from wild type and diseas. (heart) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR306536(GSM750579) 19-24nt. (ago2 brain) | SRR345199(SRX097260) source: size fractionated RNA from mouse hipp. (brain) | SRR206939(GSM723280) other. (brain) | SRR039154(GSM471931) HL1_siSrf2_smallRNAseq. (muscle) | SRR042449(GSM539841) mouse B cells activated in vitro with LPS/IL4 replicate 2 [09-002]. (b cell) | SRR306542(GSM750585) 19-24nt. (ago2 brain) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) | SRR042470(GSM539862) mouse Th17 cells [09-002]. (lymph) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................TCTGGGGCGGGAGAGCCAAAGGC.................................................................................................. | 23 | 1 | 12.00 | 12.00 | - | - | - | 7.00 | - | - | 1.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .................................................................................................................................TCTGGGGCGGGAGAGCCAAAGGCTTTTGCT........................................................................................... | 30 | 1 | 10.00 | 10.00 | - | - | 10.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................CGACGTGGTGTTCTCCATGGACCAG.......... | 25 | 1 | 10.00 | 10.00 | - | 10.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................TTGCTAAAAGATGCAGATACTCTGA....................................................................... | 25 | 1 | 6.00 | 6.00 | - | 5.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................TCTGGGGCGGGAGAGCCAAAGGCTTTTGCTAAAAGATGCAGATACTCTGAGCCGCTCTCTCTTCCCATCAG.................................................. | 71 | 1 | 6.00 | 6.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................TCTGGGGCGGGAGAGCCAAAGGT.................................................................................................. | 23 | 1 | 6.00 | 5.00 | - | - | - | - | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................TCTGGGGCGGGAGAGCCAAAGGCT................................................................................................. | 24 | 1 | 6.00 | 6.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | 2.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................TCTGGGGCGGGAGAGCCAAAGGCTT................................................................................................ | 25 | 1 | 5.00 | 5.00 | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................TCTGGGGCGGGAGAGCCAAAGG................................................................................................... | 22 | 1 | 5.00 | 5.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................GAGATCGAGAAGAACGACGT............................. | 20 | 1 | 5.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TGAGCCGCTCTCTCTTCCCATC.................................................... | 22 | 1 | 5.00 | 5.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .................................................................................................................................TCTGGGGCGGGAGAGCCAA...................................................................................................... | 19 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................TCTGGGGCGGGAGAGCCAAAGGCTTT............................................................................................... | 26 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................TGTCTGGGGCGGGAGAGCCAAAGGCTTTTGCTAAAAGATGCAGATACTCTGAGCCGCTCTCTCTTCCCATCAG.................................................. | 73 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TGAGCCGCTCTCTCTTCCCATCT................................................... | 23 | 1 | 3.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................TCTGGGGCGGGAGAGCCAAAGGCTTTTGCTAAAAGATGCAGATACTCTGAGCCGCTCTCTCTTC......................................................... | 64 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................TCTGGGGCGGGAGAGCCAAAGGCTTTTGCTAAAAGATGCAGATACTCTGAGCCGCTCTCTCTTCCCATCAGT................................................. | 72 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................GTGTTCTCCATGGACCAGACC....... | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TGAGCCGCTCTCTCTTCCCATT.................................................... | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................TCTGGGGCGGGAGAGCCAAAGGCA................................................................................................. | 24 | 1 | 2.00 | 12.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TGAGCCGCTCTCTCTTCCCAT..................................................... | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................TCTGGGGCGGGAGAGCCAAAGGCTTTT.............................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................GAGATCGAGAAGAACGAC............................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................TCTGGGGCGGGAGAGCCAAAGGCTTTTGCTAAAAGATGCAGATACTCTGAGCCGCT................................................................. | 56 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................................CCATGGACCAGACCGAAT... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................AGTGAGATCGAGAAGAACGACGTG............................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................GTGAGATCGAGAAGAACGACGTGGTGT........................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................TCTGGGGCGGGAGAGCCAAAGGCTTAT.............................................................................................. | 27 | 1 | 1.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................CAGAATTGGATTGTTCA............................................................................................................................................................................. | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................................................................................GTTCTCCATGGACCAGACCG...... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................TCTGGGGCGGGAGAGCCAAAGGG.................................................................................................. | 23 | 1 | 1.00 | 5.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................TCCCATCAGTGAGATCGAGAAGAACG................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TGAGCCGCTCTCTCTTCCCATCA................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................TCTGGGGCGGGAGAGCCAT...................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................................................................................................CGACGTGGTGTTCTCCATGGACCAGAC........ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................CTGGGGCGGGAGAGCCACC..................................................................................................... | 19 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................................................................................GAGAAGAACGACGTGGTGTTCTCCA.................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................TTGCTAAAAGATGCAGATACT........................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................CCGCTCTCTCTTCCCATCAGT................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................AGTGAGATCGAGAAGAAC.................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................TCCCATCAGTGAGATCGAGAAGAACGAC............................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................TCTGGGGCGGGAGAGCCAAAGGCTTA............................................................................................... | 26 | 1 | 1.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................AGATACTCTGAGCCGCCGGG.............................................................. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................CCGCTCTCTCTTCCCATCAGA................................................. | 21 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................CTGCCTAACACTTCTATG...................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................................................................................................GAGATCGAGAAGAACGACGTG............................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................TCTGGGGCGGGAGAGCCAAAGGCTTTTGCTAAAAGATGCAGATACTCTGAGCCGCTCTC.............................................................. | 59 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................TGAGATCGAGAAGAACGACGTGGTG......................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................TCCCATCAGTGAGATCGAGAAGAAC.................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................AAAGATGCAGATACTCTGAGCC.................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................TAACCTCACCGTCTCTGCTTCTGTGG.................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................TTGCTAAAAGATGCAGATACTCTG........................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................ATCGAGAAGAACGACGTGG........................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................GGTGTTCTCCATGGACCAGACC....... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................................ATGGACCAGACCGAAAGC. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................CGTCTCTGCTTCTGTGGGGAG.............................................................................................................................................................................................. | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................TCTGGGGCGGGAGAGCCAAAGGATT................................................................................................ | 25 | 1 | 1.00 | 5.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................TCAGTGAGATCGAGAAGAACGACGTG............................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................TCTGGGGCGGGAGAGCCAAAGGA.................................................................................................. | 23 | 1 | 1.00 | 5.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................GTCTGGGGCGGGAGAGCCAAAGGCTTTTGCTAAAAGATGCAGATACTCTGAGCCGCTCTCTCTTCCCATC.................................................... | 70 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................GCCGCTCTCTCTTCCCATCAGT................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................TGAGATCGAGAAGAACGAC............................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................TGGCTCACAGAATTGGAT................................................................................................................................................................................... | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - |
| ..................................................................................................................................CTGGGGCGGGAGAGC......................................................................................................... | 15 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - |
| ..................................................................................................................................CTGGGGCGGGAGAGCTAAG..................................................................................................... | 19 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 |
| GGAGCGAGCGAACAGCCCACAGGTTGAGGCTAACCTCACCGTCTCTGCTTCTGTGGCTCACAGAATTGGATTGTTTGTGTTTCTGCCTAACACTTCTCTGAGAGGTTTATCTACCAGGCTTCTGACATGTCTGGGGCGGGAGAGCCAAAGGCTTTTGCTAAAAGATGCAGATACTCTGAGCCGCTCTCTCTTCCCATCAGTGAGATCGAGAAGAACGACGTGGTGTTCTCCATGGACCAGACCGAATCCC ...............................................................................................................................((..(((((.((((((((....(((((((((........))))).......)))))))))))).))))).))................................................... ............................................................................................................................125........................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR346421(SRX098263) Global profiling of miRNA and the hairpin pre. (mixture) | mjTestesWT1() Testes Data. (testes) | mjLiverWT1() Liver Data. (liver) | mjLiverKO2() Liver Data. (Zcchc11 liver) | mjLiverKO1() Liver Data. (Zcchc11 liver) | GSM314557(GSM314557) ESC dgcr8 (Illumina). (ESC) | SRR346419(SRX098260) Global profiling of miRNA and the hairpin pre. (Lung) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR059769(GSM562831) Treg_Drosha. (spleen) | SRR346422(SRX098264) Global profiling of miRNA and the hairpin pre. (Muscle) | SRR032476(GSM485234) sRNA_delayed_deep_sequencing. (uterus) | mjLiverWT3() Liver Data. (liver) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR042463(GSM539855) mouse natural killer cells [09-002]. (spleen) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (ago2 liver) | SRR206942(GSM723283) other. (brain) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR346415(SRX098256) source: Testis. (Testes) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR042469(GSM539861) mouse Th2 cells [09-002]. (lymph) | mjLiverKO3() Liver Data. (Zcchc11 liver) | SRR346414(SRX098255) Global profiling of miRNA and the hairpin pre. (Spleen) | SRR014236(GSM319960) 10 dpp total. (testes) | SRR553583(SRX182789) source: Cerebellum. (Cerebellum) | Ago1IP812(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR279903(GSM689053) cell type: mouse embryonic stem cellcell line. (ESC) | SRR346420(SRX098261) Global profiling of miRNA and the hairpin pre. (Uterus) | SRR034118(GSM466728) Mili IP_Tdrd9+/- replicate1. (mili testes) | mjTestesWT3() Testes Data. (testes) | SRR306533(GSM750576) 19-24nt. (ago2 brain) | SRR032477(GSM485235) sRNA_activation_deep_sequencing. (uterus) | SRR042472(GSM539864) mouse iTreg cells [09-002]. (lymph) | SRR037926(GSM510464) e9p5_rep4. (embryo) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | Ago1IP517(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | SRR553585(SRX182791) source: Kidney. (Kidney) | SRR306540(GSM750583) 19-24nt. (ago2 brain) | GSM509276(GSM509276) small RNA cloning by length. (testes) | SRR037928(GSM510466) e7p5_rep2. (embryo) | SRR042468(GSM539860) mouse Th1 cells [09-002]. (lymph) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR042478(GSM539870) mouse lung tissue [09-002]. (lung) | GSM510454(GSM510454) newborn_rep10. (total RNA) | SRR042474(GSM539866) mouse embryonic stem cells [09-002]. (ESC) | SRR073954(GSM629280) total RNA. (blood) | SRR346416(SRX098257) Global profiling of miRNA and the hairpin pre. (Kidney) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR095855BC3(SRX039178) sequencing of miRNA from wild type and diseas. (heart) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR306536(GSM750579) 19-24nt. (ago2 brain) | SRR345199(SRX097260) source: size fractionated RNA from mouse hipp. (brain) | SRR206939(GSM723280) other. (brain) | SRR039154(GSM471931) HL1_siSrf2_smallRNAseq. (muscle) | SRR042449(GSM539841) mouse B cells activated in vitro with LPS/IL4 replicate 2 [09-002]. (b cell) | SRR306542(GSM750585) 19-24nt. (ago2 brain) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) | SRR042470(GSM539862) mouse Th17 cells [09-002]. (lymph) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................................................................TCTCTTCCCATCAGTGCGGA............................................ | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................................................................................CTCTCTTCCCATCAGTGG............................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | |
| ..........................................................................................................................................................................................................................CGTGGTGTTCTCCATGCG.............. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |