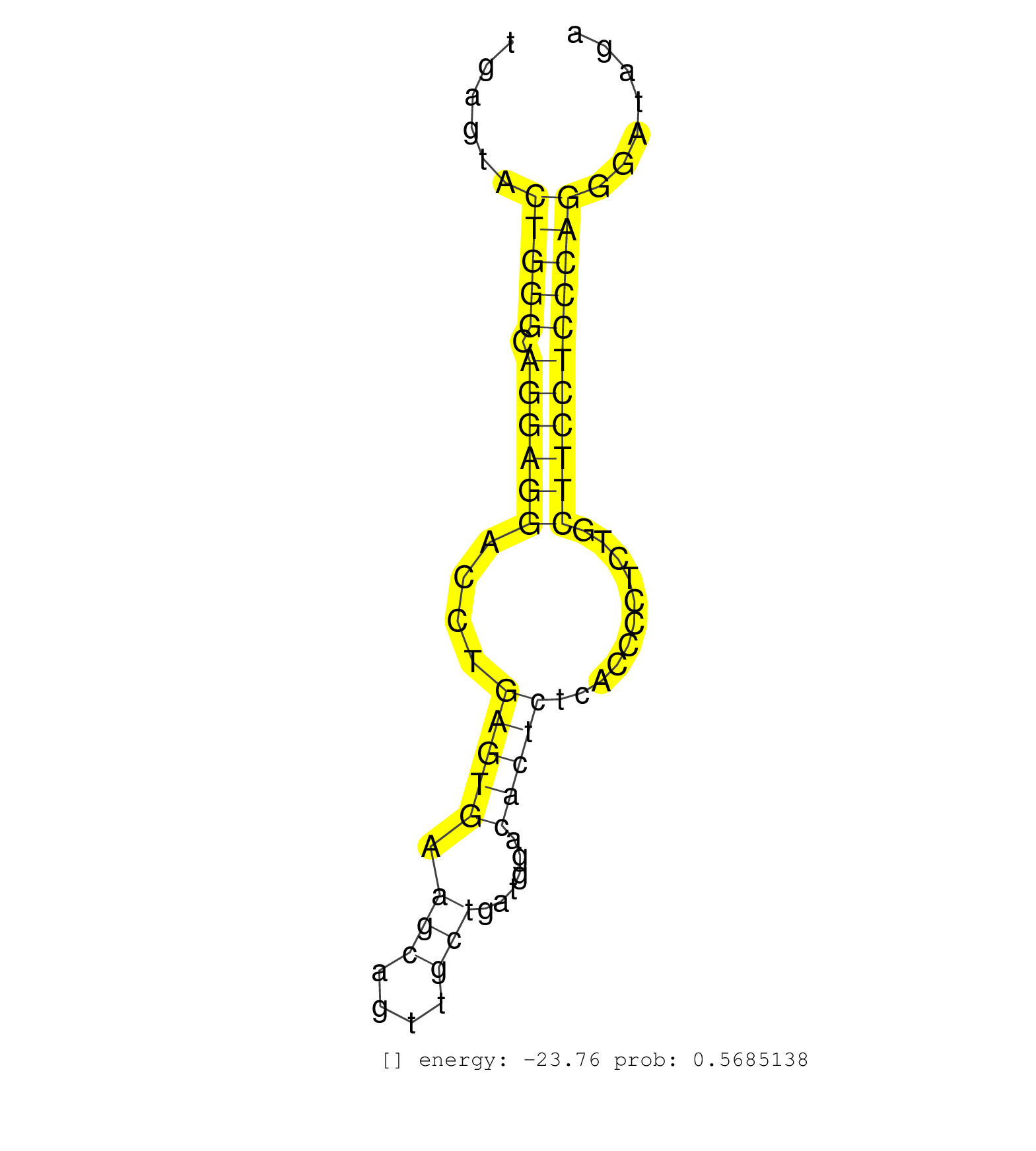
| (1) AGO.mut | (1) AGO1.ip | (5) AGO2.ip | (11) BRAIN | (1) CELL-LINE | (2) DCR.mut | (8) EMBRYO | (2) ESC | (2) FIBROBLAST | (4) LIVER | (8) OTHER | (3) OTHER.mut | (2) PIWI.ip | (1) SKIN | (3) SPLEEN | (1) TDRD1.ip | (13) TESTES | (3) TOTAL-RNA | (1) UTERUS |
| CCCTGTCTGACCTGGGCTTCTGCCTTGCCCATGGCTGCCATACAGCATGAGCTAGGCTGGTGCCTGAGAAAGCCATAGCCGAGGGCAGTGAGCACCATCATTCTGTTCCCTCGGGGACATTGTGGGATTTGAGTACTGGGCAGGAGGACCTGAGTGAAGCAGTTGCTGATGGACACTCTCACCCCTCTGCTTCCTCCCAGGGATAGACTATAAGACTACCACCATCCTCCTGGATGGACGGCGTGTCAAG .......................................................................................................................................(((((.((((((....(((((.(((....)))......)))))...........))))))))))).................................................. .................................................................................................................................130..........................................................................207......................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR685340(GSM1079784) Small RNAs (15-50 nts in length) from immorta. (dicer cell line) | SRR039184(GSM485324) shLuc.1309 (+dox). (fibroblast) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | mjLiverKO3() Liver Data. (Zcchc11 liver) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (ago2 liver) | SRR546155(SRX180174) Global profiling of miRNA and the hairpin pre. (ago2 Neuroblastoma) | SRR553583(SRX182789) source: Cerebellum. (Cerebellum) | SRR039186(GSM485326) shLuc.1309;ROSA-rtTA (+dox). (fibroblast) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | GSM510456(GSM510456) newborn_rep12. (total RNA) | SRR346424(SRX098266) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | GSM510453(GSM510453) newborn_rep9. (total RNA) | SRR391847(GSM852142) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR037924(GSM510462) e9p5_rep2. (embryo) | SRR059769(GSM562831) Treg_Drosha. (spleen) | SRR037912(GSM510449) newborn_rep5. (total RNA) | SRR553602(SRX182808) source: Sertoli cells. (Sertoli cells) | mjTestesWT2() Testes Data. (testes) | SRR553604(SRX182810) source: Spermatocytes. (Spermatocytes) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR206941(GSM723282) other. (brain) | GSM640578(GSM640578) small RNA in the liver with paternal control. (liver) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR346423(SRX098265) Global profiling of miRNA and the hairpin pre. (Brain) | SRR037930(GSM510468) e7p5_rep4. (embryo) | SRR039154(GSM471931) HL1_siSrf2_smallRNAseq. (muscle) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR553582(SRX182788) source: Brain. (Brain) | SRR206939(GSM723280) other. (brain) | SRR402766(SRX117949) source: ES PGK female cells. (ESC) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR037902(GSM510438) testes_rep3. (testes) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR073955(GSM629281) total RNA. (blood) | SRR073954(GSM629280) total RNA. (blood) | SRR306542(GSM750585) 19-24nt. (ago2 brain) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | SRR038743(GSM527278) small RNA-Seq. (dicer brain) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR306540(GSM750583) 19-24nt. (ago2 brain) | SRR306537(GSM750580) 19-24nt. (ago2 brain) | SRR037928(GSM510466) e7p5_rep2. (embryo) | Ago1IP517(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM314557(GSM314557) ESC dgcr8 (Illumina). (ESC) | SRR059772(GSM562834) CD4_Drosha. (spleen) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR553586(SRX182792) source: Testis. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR042465(GSM539857) mouse CD4+ T cells (spleen) [09-002]. (spleen) | SRR346420(SRX098261) Global profiling of miRNA and the hairpin pre. (Uterus) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................ACTGGGCAGGAGGACCTGAGTGA............................................................................................. | 23 | 1 | 15.00 | 15.00 | - | - | 2.00 | 3.00 | - | 1.00 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | 1.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ......................................................................................................................................................TGAGTGAAGCAGTTGCTGATGGACA........................................................................... | 25 | 1 | 10.00 | 10.00 | 10.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................ACCCCTCTGCTTCCTCCCAGAAT............................................... | 23 | 5.00 | 0.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................ACTGGGCAGGAGGACCTGAGTGT............................................................................................. | 23 | 1 | 5.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ......................................................................................................................................ACTGGGCAGGAGGACCTGAGT............................................................................................... | 21 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TACTGGGCAGGAGGACCTGAGTGA............................................................................................. | 24 | 1 | 4.00 | 4.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................ACTGGGCAGGAGGACCTGAGTG.............................................................................................. | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................TGAGCTAGGCTGGTGCCTGAG...................................................................................................................................................................................... | 21 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................TGGGATTTGAGTACTGGGCA............................................................................................................ | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................CCATAGCCGAGGGCAGTGAGT............................................................................................................................................................. | 21 | 2.00 | 0.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................................................................CCCCTCTGCTTCCTCCCAGT................................................. | 20 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................TGAGCTAGGCTGGTGCCTGAGAAC................................................................................................................................................................................... | 24 | 2.00 | 0.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................ACTGGGCAGGAGGACCTGA................................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................TAAGACTACCACCATCCTCCTGGATGGACGGC........ | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ...............................................................................CGAGGGCAGTGAGCAAAG......................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................TACTGGGCAGGAGGACCTGAGTGT............................................................................................. | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................GGAGGACCTGAGTGAAGCAGTTGCTGATA............................................................................... | 29 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................ACTGGGCAGGAGGACCTGAT................................................................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................................TCCTCCTGGATGGACGGCGTGTCAAGT | 27 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................TACTGGGCAGGAGGACCTGAGTA.............................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TACTGGGCAGGAGGACCTGAG................................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................TAGACTATAAGACTACCACCATCC....................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................ACTGGGCAGGAGGACCTGAGTGAA............................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................CAGGAGGACCTGAGTGAAGCAG........................................................................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................ACTGGGCAGGAGGACCTGAGCG.............................................................................................. | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................GATTTGAGTACTGGGCAGGAGGACCTGAGTGAA............................................................................................ | 33 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ...............................................TGAGCTAGGCTGGTGCCTATT...................................................................................................................................................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................ACTGGGCAGGAGGACCTGT................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................TACTGGGCAGGAGGACCTGA................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................ACCCCTCTGCTTCCTCCAAGA................................................. | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................AGCAGTTGCTGATGGCG............................................................................ | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | |
| ......................................................................................................................................................TGAGTGAAGCAGTTGCTGATGGA............................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TACTGGGCAGGAGGACCTGAGTT.............................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................CCCCTCTGCTTCCTCCCAGTTT............................................... | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................ACTGGGCAGGAGGACCTGAGGGAA............................................................................................ | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................ACTGGGCAGGAGGACCTGAGGGA............................................................................................. | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................................................................ATAGACTATAAGACTACCACCA.......................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ......................................................................................................................................ACTGGGCAGGAGGACCTGAGTGAG............................................................................................ | 24 | 1 | 1.00 | 15.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................TGAGCTAGGCTGGTGCCTGAGA..................................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................CTGGGCAGGAGGACCTGAGT............................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................ACCCCTCTGCTTCCTCCCAGA................................................. | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................................................TAAGACTACCACCATCCTCCTGGATGGACGG......... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................ACTGGGCAGGAGGACCTGAAAAA............................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TACTGGGCAGGAGGACCTGAGT............................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CCCTGTCTGACCTGGGCTTCTGCCTTGCCCATGGCTGCCATACAGCATGAGCTAGGCTGGTGCCTGAGAAAGCCATAGCCGAGGGCAGTGAGCACCATCATTCTGTTCCCTCGGGGACATTGTGGGATTTGAGTACTGGGCAGGAGGACCTGAGTGAAGCAGTTGCTGATGGACACTCTCACCCCTCTGCTTCCTCCCAGGGATAGACTATAAGACTACCACCATCCTCCTGGATGGACGGCGTGTCAAG .......................................................................................................................................(((((.((((((....(((((.(((....)))......)))))...........))))))))))).................................................. .................................................................................................................................130..........................................................................207......................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR685340(GSM1079784) Small RNAs (15-50 nts in length) from immorta. (dicer cell line) | SRR039184(GSM485324) shLuc.1309 (+dox). (fibroblast) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | mjLiverKO3() Liver Data. (Zcchc11 liver) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (ago2 liver) | SRR546155(SRX180174) Global profiling of miRNA and the hairpin pre. (ago2 Neuroblastoma) | SRR553583(SRX182789) source: Cerebellum. (Cerebellum) | SRR039186(GSM485326) shLuc.1309;ROSA-rtTA (+dox). (fibroblast) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | GSM510456(GSM510456) newborn_rep12. (total RNA) | SRR346424(SRX098266) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | GSM510453(GSM510453) newborn_rep9. (total RNA) | SRR391847(GSM852142) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR037924(GSM510462) e9p5_rep2. (embryo) | SRR059769(GSM562831) Treg_Drosha. (spleen) | SRR037912(GSM510449) newborn_rep5. (total RNA) | SRR553602(SRX182808) source: Sertoli cells. (Sertoli cells) | mjTestesWT2() Testes Data. (testes) | SRR553604(SRX182810) source: Spermatocytes. (Spermatocytes) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR206941(GSM723282) other. (brain) | GSM640578(GSM640578) small RNA in the liver with paternal control. (liver) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR346423(SRX098265) Global profiling of miRNA and the hairpin pre. (Brain) | SRR037930(GSM510468) e7p5_rep4. (embryo) | SRR039154(GSM471931) HL1_siSrf2_smallRNAseq. (muscle) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR553582(SRX182788) source: Brain. (Brain) | SRR206939(GSM723280) other. (brain) | SRR402766(SRX117949) source: ES PGK female cells. (ESC) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR037902(GSM510438) testes_rep3. (testes) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | GSM509280(GSM509280) small RNA cloning by length. (testes) | mjTestesKO8() Testes Data. (Zcchc11 testes) | SRR073955(GSM629281) total RNA. (blood) | SRR073954(GSM629280) total RNA. (blood) | SRR306542(GSM750585) 19-24nt. (ago2 brain) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | SRR038743(GSM527278) small RNA-Seq. (dicer brain) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR306540(GSM750583) 19-24nt. (ago2 brain) | SRR306537(GSM750580) 19-24nt. (ago2 brain) | SRR037928(GSM510466) e7p5_rep2. (embryo) | Ago1IP517(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | SRR034119(GSM466729) Mili IP_Tdrd9+/- replicate2. (mili testes) | GSM314557(GSM314557) ESC dgcr8 (Illumina). (ESC) | SRR059772(GSM562834) CD4_Drosha. (spleen) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR553586(SRX182792) source: Testis. (testes) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR248527(GSM733815) cell type: spermatogonial stem cell enriched . (testes) | SRR028730(GSM400967) Tdrd1-associated. (tdrd1 testes) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR042465(GSM539857) mouse CD4+ T cells (spleen) [09-002]. (spleen) | SRR346420(SRX098261) Global profiling of miRNA and the hairpin pre. (Uterus) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................................................................CTGCTTCCTCCCAGGGATAGACTATAT..................................... | 27 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | |
| .........................................................................................................................................................................TGGACACTCTCACCCCCC............................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................AAGCCATAGCCGAGGGCAGTGAGCAC........................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................TAAGACTACCACCATCGAT..................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................................................CTCTCACCCCTCTGCTACGA....................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................................................................CCTCTGCTTCCTCCCAGGGATAGACTA........................................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |