| (1) AGO.mut | (2) AGO1.ip | (1) AGO2.ip | (2) AGO3.ip | (4) BRAIN | (1) DGCR8.mut | (1) EMBRYO | (1) ESC | (1) HEART | (2) LIVER | (1) LUNG | (3) OTHER | (6) SKIN | (1) TESTES | (3) TOTAL-RNA | (1) UTERUS |
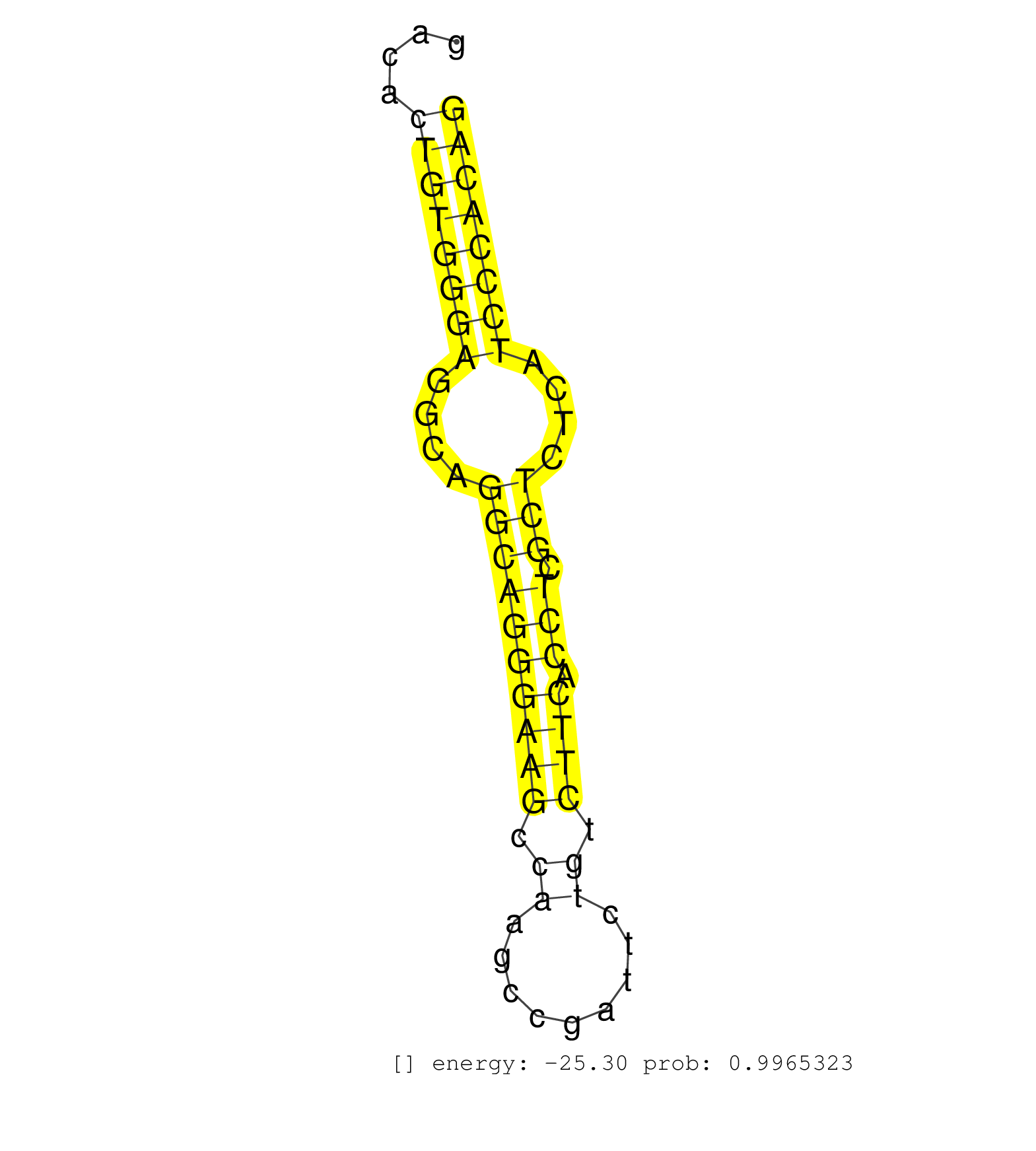
| TTTCTTTGTTCACTGGTTTGTCCTCAGTGCCTAAAACCGTGCAACACATCTTCATGGTGCTCAATAAATGCCTGCAAGTAAATGTGTAGAGGATCAGCGACTGATCCAAGGACCTGGCTGGGAAGTAACAGAGTGGACACTGTGGGAGGCAGGCAGGGAAGCCAAGCCGATTCTGTCTTCACCTCGCTCTCATCCCACAGACAGCATCACAGGCACGGCAGCAGCACCTGAGCTCGCTGCAGGATTACAT ...........................................................................................................................................((((((((....((((((((((.((.........)).)))).))).)))....)))))))).................................................. .......................................................................................................................................136.............................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | Ago1IP517(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | Ago1IP812(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | Ago3IP812(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | Ago3IP805(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | RuiDGCR8KO(Rui) DGCR8-dependent microRNA biogenesis is essent. (dgcr8 skin) | SRR346421(SRX098263) Global profiling of miRNA and the hairpin pre. (mixture) | RuiDGCR8WT(Rui) DGCR8-dependent microRNA biogenesis is essent. (skin) | GSM314557(GSM314557) ESC dgcr8 (Illumina). (ESC) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (ago2 liver) | SRR346419(SRX098260) Global profiling of miRNA and the hairpin pre. (Lung) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | GSM510452(GSM510452) newborn_rep8. (total RNA) | SRR037914(GSM510451) newborn_rep7. (total RNA) | SRR346413(SRX098254) Global profiling of miRNA and the hairpin pre. (Heart) | SRR346418(SRX098259) Global profiling of miRNA and the hairpin pre. (Liver) | SRR346422(SRX098264) Global profiling of miRNA and the hairpin pre. (Muscle) | SRR346420(SRX098261) Global profiling of miRNA and the hairpin pre. (Uterus) | SRR345206(SRX097267) source: size fractionated RNA from mouse hipp. (brain) | SRR065054(SRR065054) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR306530(GSM750573) 19-24nt. (ago2 brain) | GSM510454(GSM510454) newborn_rep10. (total RNA) | mjTestesWT3() Testes Data. (testes) | SRR345205(SRX097266) source: size fractionated RNA from mouse hipp. (brain) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................................................CTTCACCTCGCTCTCATCCCACAGT................................................. | 25 | 1 | 21.00 | 15.00 | 7.00 | 2.00 | 2.00 | 3.00 | 4.00 | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................CTTCACCTCGCTCTCATCCCACAG.................................................. | 24 | 1 | 15.00 | 15.00 | 6.00 | 5.00 | 1.00 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................CTTCACCTCGCTCTCATCCCACA................................................... | 23 | 1 | 12.00 | 12.00 | 5.00 | 5.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TTCACCTCGCTCTCATCCCACAG.................................................. | 23 | 1 | 5.00 | 5.00 | - | 2.00 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................CTTCACCTCGCTCTCATCCCAC.................................................... | 22 | 1 | 5.00 | 5.00 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TTCACCTCGCTCTCATCCCACAGT................................................. | 24 | 1 | 4.00 | 5.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - |
| .................................................................................................................................................................................TTCACCTCGCTCTCATCCCACCGT................................................. | 24 | 3.00 | 0.00 | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................................TTCACCTCGCTCTCATCCCACCGA................................................. | 24 | 2.00 | 0.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................TGTGGGAGGCAGGCAGGGAAGT........................................................................................ | 22 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................TGTGGGAGGCAGGCAGGGAAG......................................................................................... | 21 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ................................................................................................................................................................................CTTCACCTCGCTCTCATCCCACAGC................................................. | 25 | 1 | 2.00 | 15.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................CTTCACCTCGCTCTCATCCCATA................................................... | 23 | 1 | 2.00 | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................GCTGGGAAGTAACAGAGTGGACACTGTGGGAGGCAGGCAGGGAAGCCAAGCCGATTCTGTCTTCACCTCGCTCTCATCCCACAG.................................................. | 84 | 1 | 2.00 | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................CTTCACCTCGCTCTCATCCCACAGA................................................. | 25 | 1 | 2.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................TGTGGGAGGCAGGCAGGGAAGA........................................................................................ | 22 | 1 | 1.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................AGCATCACAGGCACGGCAGCAGCACCT..................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................CTTCACCTCGCTCTCATCCCACT................................................... | 23 | 1 | 1.00 | 5.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................CAGCAGCACCTGAGCTCGGTT........... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | |
| ............................................................................................................................................TGTGGGAGGCAGGCAGGGAAGC........................................................................................ | 22 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................CTTCACCTCGCTCTCATCCCAAAG.................................................. | 24 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................TGTGGGAGGCAGGCAGGGAAT......................................................................................... | 21 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................................TTCACCTCGCTCTCATCCCACCG.................................................. | 23 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................................................GGCACGGCAGCAGCACCTG.................... | 19 | 2 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................CAGCACCTGAGCTCGCTGCAGGATT.... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .................................................................................................................................................................................TTCACCTCGCTCTCATCCCACA................................................... | 22 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................CTTCACCTCGCTCTCATCCCACAA.................................................. | 24 | 1 | 1.00 | 12.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................TGTGGGAGGCAGGCAGGGAAGCC....................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................CTTCACCTCGCTCTCATCCCA..................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................CACAGGCACGGCAGCAGCACCTGAGCTCG.............. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................CTTCACCTCGCTCTCATCCTACT................................................... | 23 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................................................................................CAGCAGCACCTGAGCT................ | 16 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 |
| TTTCTTTGTTCACTGGTTTGTCCTCAGTGCCTAAAACCGTGCAACACATCTTCATGGTGCTCAATAAATGCCTGCAAGTAAATGTGTAGAGGATCAGCGACTGATCCAAGGACCTGGCTGGGAAGTAACAGAGTGGACACTGTGGGAGGCAGGCAGGGAAGCCAAGCCGATTCTGTCTTCACCTCGCTCTCATCCCACAGACAGCATCACAGGCACGGCAGCAGCACCTGAGCTCGCTGCAGGATTACAT ...........................................................................................................................................((((((((....((((((((((.((.........)).)))).))).)))....)))))))).................................................. .......................................................................................................................................136.............................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | Ago1IP517(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | Ago1IP812(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | Ago3IP812(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | Ago3IP805(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | RuiDGCR8KO(Rui) DGCR8-dependent microRNA biogenesis is essent. (dgcr8 skin) | SRR346421(SRX098263) Global profiling of miRNA and the hairpin pre. (mixture) | RuiDGCR8WT(Rui) DGCR8-dependent microRNA biogenesis is essent. (skin) | GSM314557(GSM314557) ESC dgcr8 (Illumina). (ESC) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (ago2 liver) | SRR346419(SRX098260) Global profiling of miRNA and the hairpin pre. (Lung) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | GSM510452(GSM510452) newborn_rep8. (total RNA) | SRR037914(GSM510451) newborn_rep7. (total RNA) | SRR346413(SRX098254) Global profiling of miRNA and the hairpin pre. (Heart) | SRR346418(SRX098259) Global profiling of miRNA and the hairpin pre. (Liver) | SRR346422(SRX098264) Global profiling of miRNA and the hairpin pre. (Muscle) | SRR346420(SRX098261) Global profiling of miRNA and the hairpin pre. (Uterus) | SRR345206(SRX097267) source: size fractionated RNA from mouse hipp. (brain) | SRR065054(SRR065054) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR306530(GSM750573) 19-24nt. (ago2 brain) | GSM510454(GSM510454) newborn_rep10. (total RNA) | mjTestesWT3() Testes Data. (testes) | SRR345205(SRX097266) source: size fractionated RNA from mouse hipp. (brain) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................................TCTTCACCTCGCTCTAGG......................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | |
| ...............GTTTGTCCTCAGTGCAT.......................................................................................................................................................................................................................... | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | |
| ................TTTGTCCTCAGTGCCTA......................................................................................................................................................................................................................... | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................................CCACAGACAGCATCACA....................................... | 17 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - |