| (20) AGO2.ip | (1) B-CELL | (37) BRAIN | (1) CELL-LINE | (1) DCR.mut | (2) DGCR8.mut | (5) EMBRYO | (2) ESC | (1) FIBROBLAST | (1) KIDNEY | (1) LUNG | (5) OTHER | (5) OTHER.mut | (1) OVARY | (1) PIWI.mut | (8) TESTES |
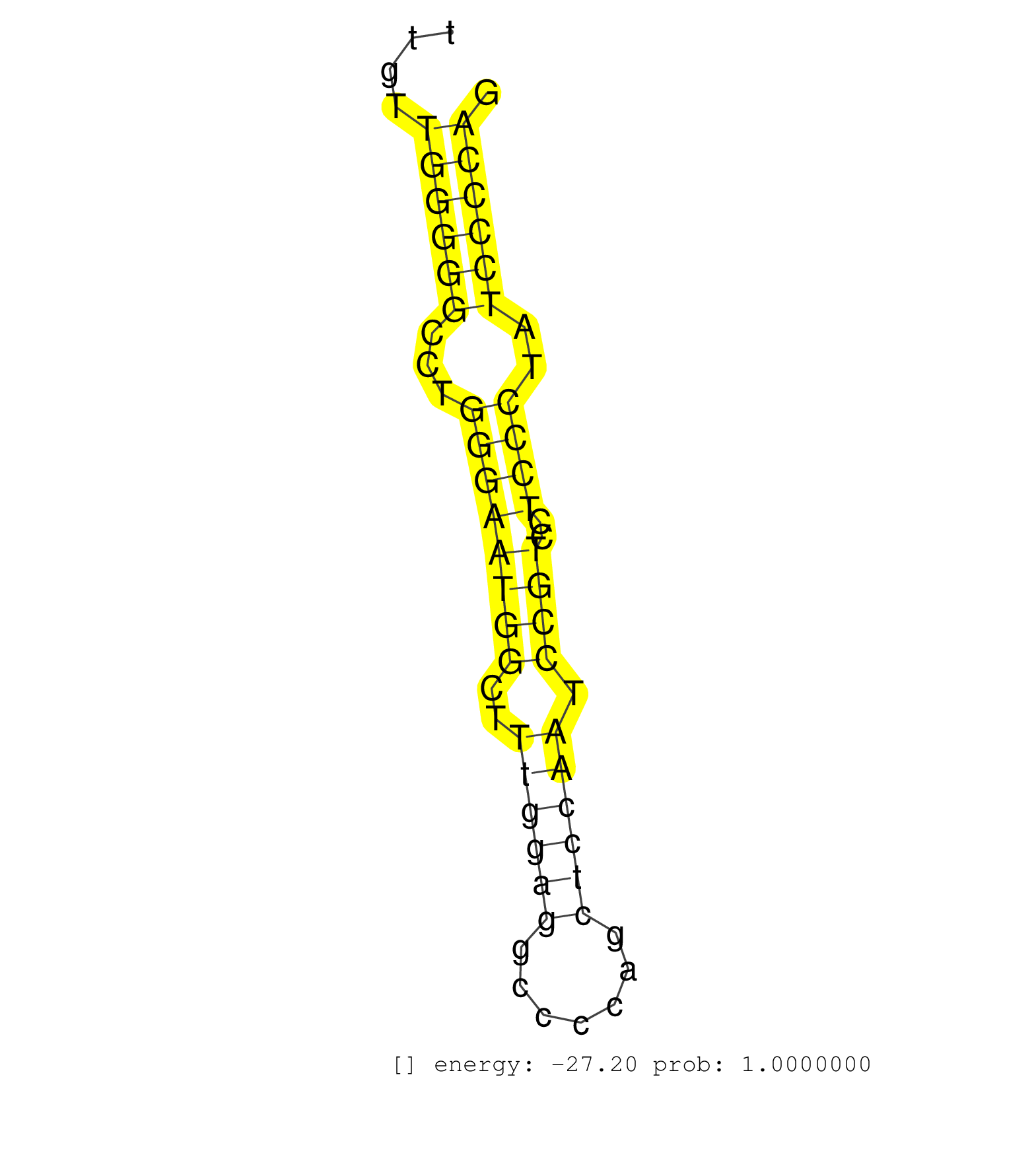
| AAACATCTGGATGGAGGGTGCTATTTAGGGGGACACGGACAAGGAACCAGGGGATACCTTTGTACCACGCTGAGAGACACTGGGCAGGATGTCGTCCCTCTTTGGGCCCCACTCTTTGTTATCTATCCAATGGCAGTATTGTTGGGGGCCTGGGAATGGCTTTGGAGGCCCCAGCTCCAATCCGTCCTCCCTATCCCCAGGACAGCTCGAGCCTCGGCCGGCCCCGTGTTCAAGGGCGTCTGTAAGCAGT ..............................................................................................................................................((((((...((((((((..((((((.......)))))).))))..))))..))))))................................................... ..........................................................................................................................................139..........................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR306530(GSM750573) 19-24nt. (ago2 brain) | SRR306531(GSM750574) 19-24nt. (ago2 brain) | SRR306529(GSM750572) 19-24nt. (ago2 brain) | SRR306542(GSM750585) 19-24nt. (ago2 brain) | SRR306533(GSM750576) 19-24nt. (ago2 brain) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | SRR306536(GSM750579) 19-24nt. (ago2 brain) | SRR553583(SRX182789) source: Cerebellum. (Cerebellum) | SRR306532(GSM750575) 19-24nt. (ago2 brain) | SRR306540(GSM750583) 19-24nt. (ago2 brain) | SRR306535(GSM750578) 19-24nt. (ago2 brain) | GSM510444(GSM510444) brain_rep5. (brain) | SRR306537(GSM750580) 19-24nt. (ago2 brain) | SRR306534(GSM750577) 19-24nt. (ago2 brain) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR546155(SRX180174) Global profiling of miRNA and the hairpin pre. (ago2 Neuroblastoma) | SRR346417(SRX098258) Global profiling of miRNA and the hairpin pre. (Brain) | SRR206941(GSM723282) other. (brain) | SRR306527(GSM750570) 19-24nt. (ago2 brain) | SRR306526(GSM750569) 19-24nt. (ago2 brain) | SRR306544(GSM750587) 19-24nt. (ago2 brain) | SRR306539(GSM750582) 19-24nt. (ago2 brain) | SRR038744(GSM527279) small RNA-Seq. (brain) | SRR306543(GSM750586) 19-24nt. (ago2 brain) | SRR206942(GSM723283) other. (brain) | SRR553582(SRX182788) source: Brain. (Brain) | SRR037929(GSM510467) e7p5_rep3. (embryo) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR038741(GSM527276) small RNA-Seq. (brain) | SRR206940(GSM723281) other. (brain) | SRR039610(GSM527274) small RNA-Seq. (brain) | SRR038745(GSM527280) small RNA-Seq. (dgcr8 brain) | SRR346421(SRX098263) Global profiling of miRNA and the hairpin pre. (mixture) | SRR346423(SRX098265) Global profiling of miRNA and the hairpin pre. (Brain) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR279905(GSM689055) cell type: mouse embryonic stem cellcell line. (ESC) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR306528(GSM750571) 19-24nt. (ago2 brain) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR037928(GSM510466) e7p5_rep2. (embryo) | SRR116846(GSM678422) Small RNA deep sequencing from mouse epididym. (epididymis) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR394083(GSM855969) background strain: C57BL6/SV129cell type: KRa. (cell line) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR346424(SRX098266) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR042450(GSM539842) mouse B cells activated in vitro with LPS/IL4 replicate 3 [09-002]. (b cell) | SRR206939(GSM723280) other. (brain) | SRR345201(SRX097262) source: size fractionated RNA from mouse hipp. (brain) | SRR037905(GSM510441) brain_rep2. (brain) | SRR037903(GSM510439) testes_rep4. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR346416(SRX098257) Global profiling of miRNA and the hairpin pre. (Kidney) | SRR038740(GSM527275) small RNA-Seq. (dicer brain) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR039183(GSM485323) shLuc.1309 (-dox). (fibroblast) | SRR346419(SRX098260) Global profiling of miRNA and the hairpin pre. (Lung) | SRR306538(GSM750581) 19-24nt. (ago2 brain) | GSM314557(GSM314557) ESC dgcr8 (Illumina). (ESC) | SRR037897(GSM510433) ovary_rep2. (ovary) | mjTestesWT3() Testes Data. (testes) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) | SRR345199(SRX097260) source: size fractionated RNA from mouse hipp. (brain) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................................AATCCGTCCTCCCTATCCCCAGT................................................. | 23 | 1 | 262.00 | 60.00 | 33.00 | 31.00 | 22.00 | 23.00 | 13.00 | 19.00 | 14.00 | 5.00 | 8.00 | 11.00 | 11.00 | 4.00 | 9.00 | 6.00 | 7.00 | 3.00 | 4.00 | - | 2.00 | 4.00 | 6.00 | 2.00 | 1.00 | 4.00 | 3.00 | - | - | 2.00 | 3.00 | 1.00 | 2.00 | 1.00 | - | 2.00 | - | - | - | 2.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................AATCCGTCCTCCCTATCCCCAG.................................................. | 22 | 1 | 60.00 | 60.00 | 9.00 | 1.00 | 3.00 | 5.00 | 8.00 | 1.00 | 2.00 | 4.00 | 5.00 | 1.00 | - | 3.00 | 2.00 | 1.00 | - | 2.00 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | 2.00 | - | 1.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................................................................................................................................................AATCCGTCCTCCCTATCCCCAGA................................................. | 23 | 1 | 26.00 | 60.00 | - | - | 1.00 | 1.00 | - | - | - | 5.00 | - | 1.00 | 2.00 | 1.00 | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | 2.00 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | 1.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................AATCCGTCCTCCCTATCCCCA................................................... | 21 | 1 | 22.00 | 22.00 | 2.00 | 1.00 | 2.00 | - | - | - | 1.00 | - | 2.00 | 1.00 | 2.00 | 4.00 | - | - | 2.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .................................................................................................................................................................................CAATCCGTCCTCCCTATCCCCAGT................................................. | 24 | 1 | 10.00 | 1.00 | - | - | - | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | 1.00 | 3.00 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................AATCCGTCCTCCCTATCCCC.................................................... | 20 | 1 | 9.00 | 9.00 | 2.00 | - | - | - | - | - | - | 2.00 | - | - | - | 1.00 | - | - | - | - | 1.00 | - | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................TCCTCCCTATCCCCAGGACAGT............................................ | 22 | 6.00 | 0.00 | 1.00 | 2.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................AATCCGTCCTCCCTATCCCCAGTT................................................ | 24 | 1 | 4.00 | 60.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................CAATCCGTCCTCCCTATCCCCA................................................... | 22 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................AATCCGTCCTCCCTATCCC..................................................... | 19 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................ATCCGTCCTCCCTATCCCCAGT................................................. | 22 | 2.00 | 0.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................AATCCGTCCTCCCTATCCCCAT.................................................. | 22 | 1 | 2.00 | 22.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................AATCCGTCCTCCCTATCCCCCGT................................................. | 23 | 1 | 2.00 | 9.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................AATCCGTCCTCCCTATCCCCAGC................................................. | 23 | 1 | 2.00 | 60.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................ATCCGTCCTCCCTATCCCCA................................................... | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TTGGGGGCCTGGGAATGGCTTTG...................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................CAATCCGTCCTCCCTATCCCCAG.................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................GTCCTCCCTATCCCCAGT................................................. | 18 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....ATCTGGATGGAGGGTGAA.................................................................................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................................................GCCTCGGCCGGCCCCGG....................... | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................TTGGGGGCCTGGGAATGGCTT........................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................GTCCTCCCTATCCCCAGA................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................TGGGGGCCTGGGAATGGCTTTGGAGGCCCCAGCTCCAATCCGTCCTCCCTATCCCCAGT................................................. | 59 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................................CAATCCGTCCTCCCTATCCCCAGA................................................. | 24 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................CAATCCGTCCTCCCTATCCCCCGT................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................GGGGCCTGGGAATGGGG......................................................................................... | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................AATCCGTCCTCCCTATCCA..................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................................................................GAGCCTCGGCCGGCCCCGGA...................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................................................................................................CAAGGGCGTCTGTAAGC... | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .....TCTGGATGGAGGGTGCATAT................................................................................................................................................................................................................................. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................TGGGAATGGCTTTGGCG................................................................................... | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | |
| ....................................................................................................................................................................................................................................TTCAAGGGCGTCTGTAAGC... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................ATCTATCCAATGGCAGT................................................................................................................. | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................GGACAAGGAACCAGGGGATA.................................................................................................................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................CGTCCTCCCTATCCCCAGT................................................. | 19 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................AATCCGTCCTCCCTATCCCCAGAT................................................ | 24 | 1 | 1.00 | 60.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ..........................AGGGGGACACGGACAAGGAACC.......................................................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................CAATCCGTCCTCCCTATCCCC.................................................... | 21 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................AATCCGTCCTCCCTATCCCCAGTA................................................ | 24 | 1 | 1.00 | 60.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................AATCCGTCCTCCCTATCCCCATT................................................. | 23 | 1 | 1.00 | 22.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................CCGTCCTCCCTATCCCCAGT................................................. | 20 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................................................................................CGAGCCTCGGCCGGCGCC......................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | |
| .............................................................................................................................................TTGGGGGCCTGGGAATGGCTTTGGAGGCCCCAGCTCCAATCCGTCCTCCCTATCCCCAGA................................................. | 60 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................................................................................GTGTTCAAGGGCGTCT......... | 16 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................CCGGCCCCGTGTT.................... | 13 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 |
| ..................................................................................................................................................................................AATCCGTCCTCCCT.......................................................... | 14 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AAACATCTGGATGGAGGGTGCTATTTAGGGGGACACGGACAAGGAACCAGGGGATACCTTTGTACCACGCTGAGAGACACTGGGCAGGATGTCGTCCCTCTTTGGGCCCCACTCTTTGTTATCTATCCAATGGCAGTATTGTTGGGGGCCTGGGAATGGCTTTGGAGGCCCCAGCTCCAATCCGTCCTCCCTATCCCCAGGACAGCTCGAGCCTCGGCCGGCCCCGTGTTCAAGGGCGTCTGTAAGCAGT ..............................................................................................................................................((((((...((((((((..((((((.......)))))).))))..))))..))))))................................................... ..........................................................................................................................................139..........................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR306530(GSM750573) 19-24nt. (ago2 brain) | SRR306531(GSM750574) 19-24nt. (ago2 brain) | SRR306529(GSM750572) 19-24nt. (ago2 brain) | SRR306542(GSM750585) 19-24nt. (ago2 brain) | SRR306533(GSM750576) 19-24nt. (ago2 brain) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | SRR306536(GSM750579) 19-24nt. (ago2 brain) | SRR553583(SRX182789) source: Cerebellum. (Cerebellum) | SRR306532(GSM750575) 19-24nt. (ago2 brain) | SRR306540(GSM750583) 19-24nt. (ago2 brain) | SRR306535(GSM750578) 19-24nt. (ago2 brain) | GSM510444(GSM510444) brain_rep5. (brain) | SRR306537(GSM750580) 19-24nt. (ago2 brain) | SRR306534(GSM750577) 19-24nt. (ago2 brain) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR546155(SRX180174) Global profiling of miRNA and the hairpin pre. (ago2 Neuroblastoma) | SRR346417(SRX098258) Global profiling of miRNA and the hairpin pre. (Brain) | SRR206941(GSM723282) other. (brain) | SRR306527(GSM750570) 19-24nt. (ago2 brain) | SRR306526(GSM750569) 19-24nt. (ago2 brain) | SRR306544(GSM750587) 19-24nt. (ago2 brain) | SRR306539(GSM750582) 19-24nt. (ago2 brain) | SRR038744(GSM527279) small RNA-Seq. (brain) | SRR306543(GSM750586) 19-24nt. (ago2 brain) | SRR206942(GSM723283) other. (brain) | SRR553582(SRX182788) source: Brain. (Brain) | SRR037929(GSM510467) e7p5_rep3. (embryo) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR038741(GSM527276) small RNA-Seq. (brain) | SRR206940(GSM723281) other. (brain) | SRR039610(GSM527274) small RNA-Seq. (brain) | SRR038745(GSM527280) small RNA-Seq. (dgcr8 brain) | SRR346421(SRX098263) Global profiling of miRNA and the hairpin pre. (mixture) | SRR346423(SRX098265) Global profiling of miRNA and the hairpin pre. (Brain) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR279905(GSM689055) cell type: mouse embryonic stem cellcell line. (ESC) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR306528(GSM750571) 19-24nt. (ago2 brain) | mjTestesKO6() Testes Data. (Zcchc11 testes) | SRR037928(GSM510466) e7p5_rep2. (embryo) | SRR116846(GSM678422) Small RNA deep sequencing from mouse epididym. (epididymis) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR394083(GSM855969) background strain: C57BL6/SV129cell type: KRa. (cell line) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR346424(SRX098266) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR042450(GSM539842) mouse B cells activated in vitro with LPS/IL4 replicate 3 [09-002]. (b cell) | SRR206939(GSM723280) other. (brain) | SRR345201(SRX097262) source: size fractionated RNA from mouse hipp. (brain) | SRR037905(GSM510441) brain_rep2. (brain) | SRR037903(GSM510439) testes_rep4. (testes) | GSM509277(GSM509277) small RNA cloning by length. (piwi testes) | SRR346416(SRX098257) Global profiling of miRNA and the hairpin pre. (Kidney) | SRR038740(GSM527275) small RNA-Seq. (dicer brain) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR039183(GSM485323) shLuc.1309 (-dox). (fibroblast) | SRR346419(SRX098260) Global profiling of miRNA and the hairpin pre. (Lung) | SRR306538(GSM750581) 19-24nt. (ago2 brain) | GSM314557(GSM314557) ESC dgcr8 (Illumina). (ESC) | SRR037897(GSM510433) ovary_rep2. (ovary) | mjTestesWT3() Testes Data. (testes) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) | SRR345199(SRX097260) source: size fractionated RNA from mouse hipp. (brain) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................................GGCCCCAGCTCCAATACA.................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................................................GCCCCAGCTCCAATCCG.................................................................. | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................AGGCCCCAGCTCCAATAA................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |