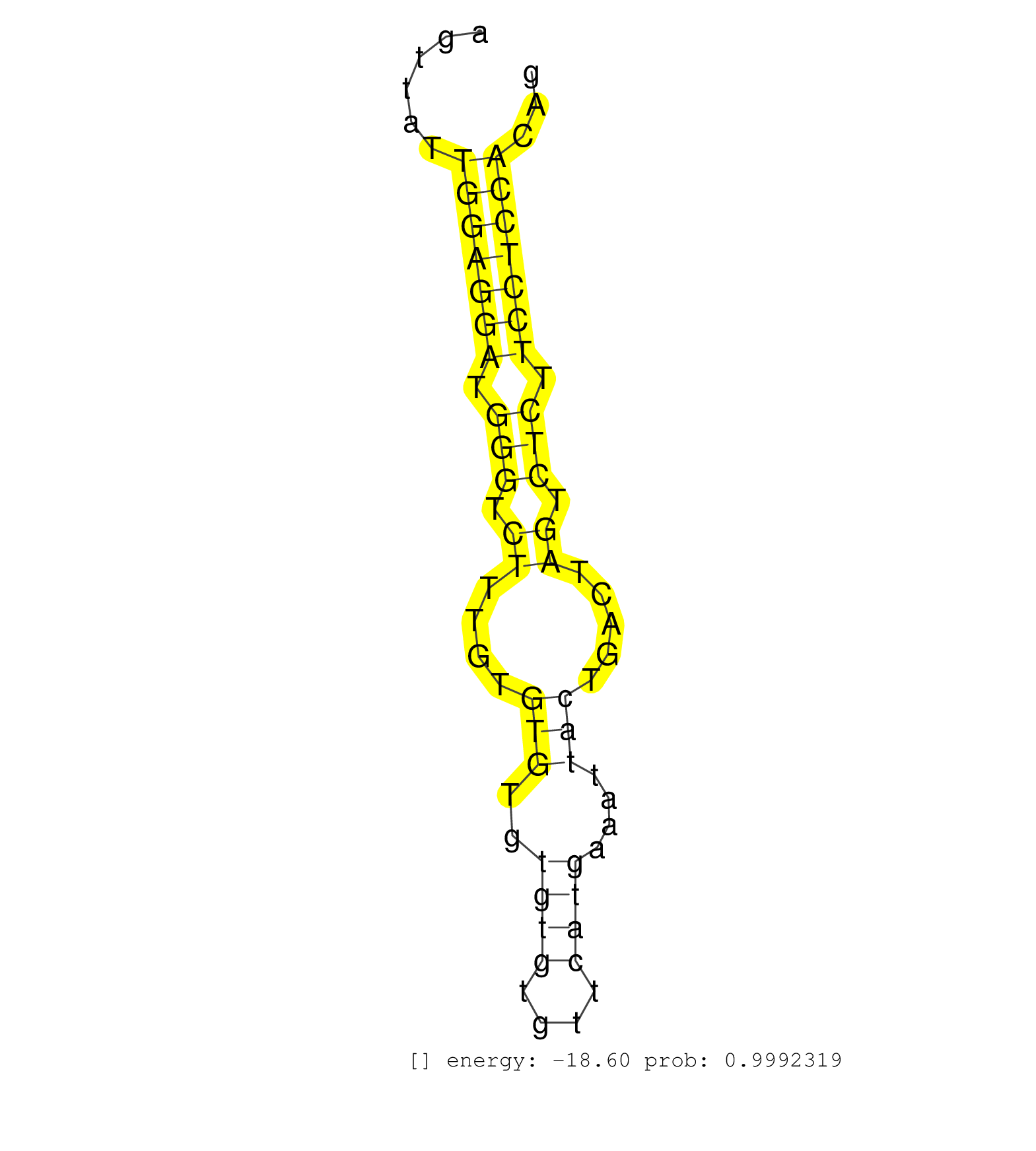
| (1) AGO.mut | (2) AGO1.ip | (3) AGO2.ip | (2) AGO3.ip | (2) BRAIN | (2) EMBRYO | (2) ESC | (2) LIVER | (6) OTHER | (7) SKIN |
| ATGGCACTGCTGCTACTGGTCATGAAGAGGCCCAAACTGCCTCACTGGTTTAAGTTACTTGGGCAAGATGCCCAAGTTCAGCCTCCTTTATTATAGATAATTTGGAGTGGCTCTACAAGGCTTACAGATCAGTTATTGGAGGATGGGTCTTTGTGTGTGTGTGTGTTCATGAAATTACTGACTAGTCTCTTCCTCCACAGGAAGAGAACAGGCCAGTAGGTGGGTTTTCTCTTCGTGGTTCCCTTGTGTC ........................................................................................................................................(((((((.(((.((....(((..((((....))))....))).....)).))).)))))))..................................................... ..................................................................................................................................131..................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | Ago2IP517(Rui) Quantitative functions of Argonaute proteins . (ago2 skin) | Ago3IP812(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | Ago1IP517(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | Ago1IP812(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | Ago3IP805(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (ago2 liver) | SRR402760(SRX117943) source: ES E14 male cells. (ESC) | SRR073955(GSM629281) total RNA. (blood) | SRR037907(GSM510443) brain_rep4. (brain) | SRR345205(SRX097266) source: size fractionated RNA from mouse hipp. (brain) | SRR546155(SRX180174) Global profiling of miRNA and the hairpin pre. (ago2 Neuroblastoma) | SRR039153(GSM471930) HL1_siSrf1_smallRNAseq. (muscle) | GSM640582(GSM640582) small RNA in the liver with paternal control . (liver) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | Ago2IP504(Rui) Quantitative functions of Argonaute proteins . (ago2 skin) | SRR073954(GSM629280) total RNA. (blood) | RuiDGCR8WT(Rui) DGCR8-dependent microRNA biogenesis is essent. (skin) | SRR553583(SRX182789) source: Cerebellum. (Cerebellum) | SRR042484(GSM539876) mouse salivary gland tissue [09-002]. (salivary gland) | GSM314553(GSM314553) ESC dcr (Illumina). (ESC) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................TTGGAGGATGGGTCTTTGTGTGT............................................................................................ | 23 | 1 | 76.00 | 76.00 | 27.00 | 11.00 | 11.00 | 8.00 | 9.00 | 2.00 | - | 2.00 | 1.00 | - | 1.00 | - | - | 1.00 | 1.00 | - | 1.00 | 1.00 | - | - | - |
| ..................................................................................................................................................................................TGACTAGTCTCTTCCTCCACA................................................... | 21 | 1 | 3.00 | 3.00 | - | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................TTGGAGGATGGGTCTTTGTGT.............................................................................................. | 21 | 1 | 3.00 | 3.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ........................................................................................................................................TGGAGGATGGGTCTTTGTGTGT............................................................................................ | 22 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................TTGGAGGATGGGTCTTTGTGTGA............................................................................................ | 23 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................TTGGAGGATGGGTCTTTGTGTG............................................................................................. | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................ATTGGAGGATGGGTCTTTGA................................................................................................ | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | |
| .......................................................................................................................................TTGGAGGATGGGTCTTTGTGTGAT........................................................................................... | 24 | 1 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..................................................................................................................................AGTTATTGGAGGATGGGA...................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................ATTGGAGGATGGGTCTTTGTGTGT............................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................TTGGAGGATGGGTCTTTGGGGA............................................................................................. | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................TTGGAGGATGGGTCTTTGTGTGTAT.......................................................................................... | 25 | 1 | 1.00 | 76.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................TTGGAGGATGGGTCTTTGTGTT............................................................................................. | 22 | 1 | 1.00 | 3.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................TGGAGGATGGGTCTTTGTGTGTGT.......................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ATGGCACTGCTGCTACTGGTCATGAAGAGGCCCAAACTGCCTCACTGGTTTAAGTTACTTGGGCAAGATGCCCAAGTTCAGCCTCCTTTATTATAGATAATTTGGAGTGGCTCTACAAGGCTTACAGATCAGTTATTGGAGGATGGGTCTTTGTGTGTGTGTGTGTTCATGAAATTACTGACTAGTCTCTTCCTCCACAGGAAGAGAACAGGCCAGTAGGTGGGTTTTCTCTTCGTGGTTCCCTTGTGTC ........................................................................................................................................(((((((.(((.((....(((..((((....))))....))).....)).))).)))))))..................................................... ..................................................................................................................................131..................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | Ago2IP517(Rui) Quantitative functions of Argonaute proteins . (ago2 skin) | Ago3IP812(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | Ago1IP517(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | Ago1IP812(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | Ago3IP805(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (ago2 liver) | SRR402760(SRX117943) source: ES E14 male cells. (ESC) | SRR073955(GSM629281) total RNA. (blood) | SRR037907(GSM510443) brain_rep4. (brain) | SRR345205(SRX097266) source: size fractionated RNA from mouse hipp. (brain) | SRR546155(SRX180174) Global profiling of miRNA and the hairpin pre. (ago2 Neuroblastoma) | SRR039153(GSM471930) HL1_siSrf1_smallRNAseq. (muscle) | GSM640582(GSM640582) small RNA in the liver with paternal control . (liver) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | Ago2IP504(Rui) Quantitative functions of Argonaute proteins . (ago2 skin) | SRR073954(GSM629280) total RNA. (blood) | RuiDGCR8WT(Rui) DGCR8-dependent microRNA biogenesis is essent. (skin) | SRR553583(SRX182789) source: Cerebellum. (Cerebellum) | SRR042484(GSM539876) mouse salivary gland tissue [09-002]. (salivary gland) | GSM314553(GSM314553) ESC dcr (Illumina). (ESC) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................................................................................GTTTTCTCTTCGTGG............ | 15 | 4 | 2.00 | 2.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................TGGAGTGGCTCTACAAT................................................................................................................................... | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................................................................GTCTCTTCCTCCACAGG................................................. | 17 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |