| (1) AGO.mut | (11) AGO2.ip | (1) AGO3.ip | (16) BRAIN | (4) CELL-LINE | (1) DCR.mut | (14) EMBRYO | (8) ESC | (6) FIBROBLAST | (1) HEART | (5) LIVER | (1) LUNG | (1) LYMPH | (6) OTHER | (3) OTHER.mut | (1) PANCREAS | (2) PIWI.ip | (1) SKIN | (4) SPLEEN | (9) TESTES | (2) THYMUS |
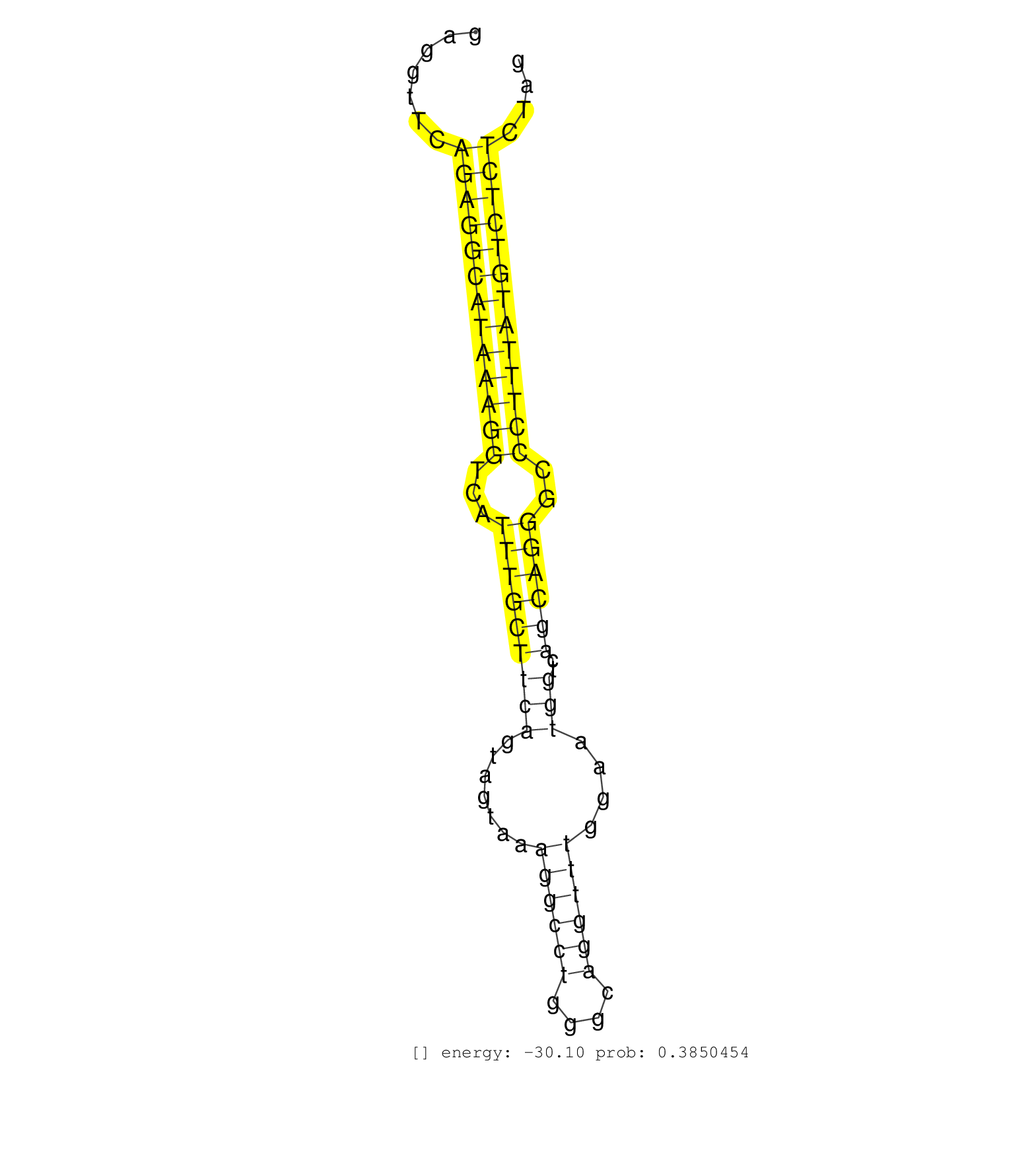
| GGATCTAGAAATAATTTCGAGACTCTAGCTCAGCCACAGCACAGACTGACTGAACCATATAAACAGGTTTTCCCTTGGTTTATTCCTCTGAAGATTTTGAGCTGGTGCAGTGAGGTTCAGAGGCATAAAGGTCATTTGCTTCAGTAGTAAAGGCCTGGGCAGGTTTGGAATGGTCAGCAGGGCCCTTTATGTCTCTCTAGGGTCGTCCCGGATGGCACATAGAGTGTTCTGCTATGGCAGGCACGCTCCT ......................................................................................................................(((((((((((((...(((((((((.......((((((....))))))....)))..))))))..)))))))))))))...................................................... ...............................................................................................................112.....................................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR391847(GSM852142) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR279903(GSM689053) cell type: mouse embryonic stem cellcell line. (ESC) | SRR402761(SRX117944) source: ES E14 male cells. (ESC) | SRR039184(GSM485324) shLuc.1309 (+dox). (fibroblast) | SRR402766(SRX117949) source: ES PGK female cells. (ESC) | SRR306544(GSM750587) 19-24nt. (ago2 brain) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR039185(GSM485325) shLuc.1309;ROSA-rtTA (-dox). (fibroblast) | SRR059769(GSM562831) Treg_Drosha. (spleen) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR306542(GSM750585) 19-24nt. (ago2 brain) | GSM314557(GSM314557) ESC dgcr8 (Illumina). (ESC) | SRR279904(GSM689054) cell type: mouse embryonic stem cellcell line. (ESC) | SRR073954(GSM629280) total RNA. (blood) | SRR546155(SRX180174) Global profiling of miRNA and the hairpin pre. (ago2 Neuroblastoma) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR039183(GSM485323) shLuc.1309 (-dox). (fibroblast) | SRR037928(GSM510466) e7p5_rep2. (embryo) | SRR059766(GSM562828) DN3_Drosha. (thymus) | SRR306537(GSM750580) 19-24nt. (ago2 brain) | SRR279906(GSM689056) cell type: mouse embryonic stem cellcell line. (ESC) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (ago2 liver) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR059765(GSM562827) DN3_control. (thymus) | SRR037920(GSM510458) e12p5_rep2. (embryo) | SRR037921(GSM510459) e12p5_rep3. (embryo) | GSM510444(GSM510444) brain_rep5. (brain) | SRR390299(GSM849857) cell line: NIH-3T3cell type: fibroblastip ant. (ago2 fibroblast) | SRR685339(GSM1079783) Small RNAs (15-50 nts in length) from immorta. (cell line) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR073955(GSM629281) total RNA. (blood) | SRR039152(GSM471929) HL1_siNon_smallRNAseq. (muscle) | SRR023852(GSM307161) ZHBT-c424hsmallrna_rep1. (cell line) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR038743(GSM527278) small RNA-Seq. (dicer brain) | SRR306534(GSM750577) 19-24nt. (ago2 brain) | SRR346419(SRX098260) Global profiling of miRNA and the hairpin pre. (Lung) | GSM640581(GSM640581) small RNA in the liver with paternal control. (liver) | SRR346414(SRX098255) Global profiling of miRNA and the hairpin pre. (Spleen) | SRR037930(GSM510468) e7p5_rep4. (embryo) | SRR039186(GSM485326) shLuc.1309;ROSA-rtTA (+dox). (fibroblast) | GSM640580(GSM640580) small RNA in the liver with paternal Low pro. (liver) | Ago3IP812(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | SRR037927(GSM510465) e7p5_rep1. (embryo) | SRR346425(SRX098267) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR023851(GSM307160) ZHBT-c412hsmallrna_rep1. (cell line) | SRR059770(GSM562832) Treg_Dicer. (spleen) | SRR023850(GSM307159) ZHBT-c40hsmallrna_rep1. (cell line) | SRR042474(GSM539866) mouse embryonic stem cells [09-002]. (ESC) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR042475(GSM539867) mouse embryonic fibroblast cells [09-002]. (fibroblast) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR306531(GSM750574) 19-24nt. (ago2 brain) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR042471(GSM539863) mouse TFHS cells [09-002]. (spleen) | SRR346421(SRX098263) Global profiling of miRNA and the hairpin pre. (mixture) | SRR042477(GSM539869) mouse brain tissue [09-002]. (brain) | SRR042470(GSM539862) mouse Th17 cells [09-002]. (lymph) | SRR077864(GSM637801) 18-30 nt small RNAs. (liver) | GSM475281(GSM475281) total RNA. (testes) | GSM361398(GSM361398) CGNP_P6_p53--_Ink4c--_rep1. (brain) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR042481(GSM539873) mouse pancreatic tissue [09-002]. (pancreas) | SRR306527(GSM750570) 19-24nt. (ago2 brain) | SRR306543(GSM750586) 19-24nt. (ago2 brain) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR306530(GSM750573) 19-24nt. (ago2 brain) | SRR038741(GSM527276) small RNA-Seq. (brain) | SRR346413(SRX098254) Global profiling of miRNA and the hairpin pre. (Heart) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................TCAGAGGCATAAAGGTCATTTGCT.............................................................................................................. | 24 | 1 | 116.00 | 116.00 | 24.00 | 21.00 | 12.00 | 9.00 | 6.00 | 11.00 | 9.00 | 4.00 | 4.00 | 2.00 | - | 1.00 | - | - | 2.00 | 1.00 | - | - | - | 1.00 | - | - | 1.00 | - | 2.00 | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................CAGAGGCATAAAGGTCATTTGCT.............................................................................................................. | 23 | 1 | 76.00 | 76.00 | 10.00 | 18.00 | 15.00 | 4.00 | 1.00 | 1.00 | - | 1.00 | 2.00 | - | - | - | - | 2.00 | 1.00 | - | 1.00 | - | 2.00 | 3.00 | 1.00 | - | - | 1.00 | - | - | - | - | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TCAGAGGCATAAAGGTCATTTGC............................................................................................................... | 23 | 1 | 47.00 | 47.00 | 6.00 | 10.00 | 7.00 | 1.00 | 1.00 | 1.00 | 1.00 | - | 1.00 | 3.00 | 1.00 | - | 1.00 | 1.00 | 1.00 | - | - | 2.00 | 3.00 | - | - | 1.00 | - | - | - | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................TTCAGAGGCATAAAGGTCATTTGCT.............................................................................................................. | 25 | 1 | 31.00 | 31.00 | 4.00 | 5.00 | 2.00 | 3.00 | - | 2.00 | 4.00 | 2.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................TTCAGAGGCATAAAGGTCATTTGC............................................................................................................... | 24 | 1 | 28.00 | 28.00 | 11.00 | 3.00 | 5.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ...................................................................................................................TTCAGAGGCATAAAGGTCATTTG................................................................................................................ | 23 | 1 | 23.00 | 23.00 | 4.00 | 4.00 | - | - | 2.00 | - | - | 1.00 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | 2.00 | - | - | - | - | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................TTCAGAGGCATAAAGGTCATTT................................................................................................................. | 22 | 1 | 20.00 | 20.00 | 4.00 | 1.00 | 5.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TCAGAGGCATAAAGGTCATTT................................................................................................................. | 21 | 1 | 13.00 | 13.00 | 3.00 | 2.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TCAGAGGCATAAAGGTCATTTG................................................................................................................ | 22 | 1 | 11.00 | 11.00 | 3.00 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................TTCAGAGGCATAAAGGTCATTTGT............................................................................................................... | 24 | 1 | 10.00 | 23.00 | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..ATCTAGAAATAATTTTGT...................................................................................................................................................................................................................................... | 18 | 6.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................CAGAGGCATAAAGGTCATTTGC............................................................................................................... | 22 | 1 | 5.00 | 5.00 | 1.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................CAGGGCCCTTTATGTCTCTCT.................................................... | 21 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................AGAGGCATAAAGGTCATTTGCT.............................................................................................................. | 22 | 1 | 3.00 | 3.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TCAGAGGCATAAAGGTCATT.................................................................................................................. | 20 | 1 | 3.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................CAGGGCCCTTTATGTCTCTCTAG.................................................. | 23 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................CAGGGCCCTTTATGTCTCTCCAT.................................................. | 23 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................CAGAGGCATAAAGGTCATTTG................................................................................................................ | 21 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................CAGAGGCATAAAGGTCATTTGCTC............................................................................................................. | 24 | 1 | 2.00 | 76.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..ATCTAGAAATAATTTTGTA..................................................................................................................................................................................................................................... | 19 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................TCAGAGGCATAAAGGTCATTTGT............................................................................................................... | 23 | 1 | 2.00 | 11.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................TTCAGAGGCATAAAGGTCATTTGCTT............................................................................................................. | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................CAGAGGCATAAAGGTCATTT................................................................................................................. | 20 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .....................................................................................................................CAGAGGCATAAAGGTCATTTGCTT............................................................................................................. | 24 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TCAGAGGCATAAAGGTCATTTGCTT............................................................................................................. | 25 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................CAGGGCCCTTTATGTCTC....................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................CAGGGCCCTTTATGTCTCTCCA................................................... | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................TCAGAGGCATAAAGGTCATTTGCA.............................................................................................................. | 24 | 1 | 1.00 | 47.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................CAGGGCCCTTTATGTCTCTCTA................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................AGAGTGTTCTGCTATGGCAGGCAC...... | 24 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................................ATGGCAGGCACGCTCCT | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........AATAATTTCGAGACTCTAGCTCAGCCACAGCACAGACTGACTGAACCATATAAA........................................................................................................................................................................................... | 54 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................GGTGCAGTGAGGTTCGGG................................................................................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................................TGGGCAGGTTTGGAATGGTC........................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ............................................................................................................................ATAAAGGTCATTTGCTAA............................................................................................................ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................AGCTGGTGCAGTGAGTTCT.................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................AGGGCCCTTTATGTCTCTCTAG.................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................ATGTCTCTCTAGGGTGTG............................................ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................................CAGGGCCCTTTATGTCTCTCA.................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................AGAGGCATAAAGGTCATTTG................................................................................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................AGCTGGTGCAGTGAGCT...................................................................................................................................... | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | |
| ....................................................................................................................TCAGAGGCATAAAGGTCTTTT................................................................................................................. | 21 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................TTCAGAGGCATAAAGGTCATTTGTT.............................................................................................................. | 25 | 1 | 1.00 | 23.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................TCTCTCTAGGGTCGTTCT......................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................AGCTGGTGCAGTGAGCCAG.................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | |
| ......................................................................................................................................................................................................AGGGTCGTCCCGGATGGCA................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................TTCAGAGGCATAAAGGTCATTC................................................................................................................. | 22 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................CATAAAGGTCATTTGCT.............................................................................................................. | 17 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - |
| ....................................................................................................................TCAGAGGCATAAAGGTCATTTAC............................................................................................................... | 23 | 1 | 1.00 | 13.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............AATTTCGAGACTCTAGCT............................................................................................................................................................................................................................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................GCATAAAGGTCATTTGCT.............................................................................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................CCCTTTATGTCTCTCTAG.................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TCAGAGGCATAAAGGTCACTT................................................................................................................. | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................................CAGGGCCCTTTATGTCTCTCTAGTA................................................ | 25 | 1 | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................AGGGCCCTTTATGTCTCTCACA.................................................. | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................CAGAGGCATAAAGGTCATT.................................................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................CATAAAGGTCATTTGC............................................................................................................... | 16 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| ....................................................................................................................TCAGAGGCATAAAGG....................................................................................................................... | 15 | 7 | 0.43 | 0.43 | 0.14 | 0.29 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| GGATCTAGAAATAATTTCGAGACTCTAGCTCAGCCACAGCACAGACTGACTGAACCATATAAACAGGTTTTCCCTTGGTTTATTCCTCTGAAGATTTTGAGCTGGTGCAGTGAGGTTCAGAGGCATAAAGGTCATTTGCTTCAGTAGTAAAGGCCTGGGCAGGTTTGGAATGGTCAGCAGGGCCCTTTATGTCTCTCTAGGGTCGTCCCGGATGGCACATAGAGTGTTCTGCTATGGCAGGCACGCTCCT ......................................................................................................................(((((((((((((...(((((((((.......((((((....))))))....)))..))))))..)))))))))))))...................................................... ...............................................................................................................112.....................................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR391847(GSM852142) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR279903(GSM689053) cell type: mouse embryonic stem cellcell line. (ESC) | SRR402761(SRX117944) source: ES E14 male cells. (ESC) | SRR039184(GSM485324) shLuc.1309 (+dox). (fibroblast) | SRR402766(SRX117949) source: ES PGK female cells. (ESC) | SRR306544(GSM750587) 19-24nt. (ago2 brain) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR029043(GSM433295) 18.5dpc_homo_tdrd1-KO. (tdrd1 testes) | SRR039185(GSM485325) shLuc.1309;ROSA-rtTA (-dox). (fibroblast) | SRR059769(GSM562831) Treg_Drosha. (spleen) | SRR029036(GSM433288) 18dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR306542(GSM750585) 19-24nt. (ago2 brain) | GSM314557(GSM314557) ESC dgcr8 (Illumina). (ESC) | SRR279904(GSM689054) cell type: mouse embryonic stem cellcell line. (ESC) | SRR073954(GSM629280) total RNA. (blood) | SRR546155(SRX180174) Global profiling of miRNA and the hairpin pre. (ago2 Neuroblastoma) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR039183(GSM485323) shLuc.1309 (-dox). (fibroblast) | SRR037928(GSM510466) e7p5_rep2. (embryo) | SRR059766(GSM562828) DN3_Drosha. (thymus) | SRR306537(GSM750580) 19-24nt. (ago2 brain) | SRR279906(GSM689056) cell type: mouse embryonic stem cellcell line. (ESC) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (ago2 liver) | SRR014232(GSM319956) 16.5 dpc MILI. (mili testes) | SRR059765(GSM562827) DN3_control. (thymus) | SRR037920(GSM510458) e12p5_rep2. (embryo) | SRR037921(GSM510459) e12p5_rep3. (embryo) | GSM510444(GSM510444) brain_rep5. (brain) | SRR390299(GSM849857) cell line: NIH-3T3cell type: fibroblastip ant. (ago2 fibroblast) | SRR685339(GSM1079783) Small RNAs (15-50 nts in length) from immorta. (cell line) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR073955(GSM629281) total RNA. (blood) | SRR039152(GSM471929) HL1_siNon_smallRNAseq. (muscle) | SRR023852(GSM307161) ZHBT-c424hsmallrna_rep1. (cell line) | SRR069811(GSM610967) small RNA sequencing; sample 3. (testes) | SRR038743(GSM527278) small RNA-Seq. (dicer brain) | SRR306534(GSM750577) 19-24nt. (ago2 brain) | SRR346419(SRX098260) Global profiling of miRNA and the hairpin pre. (Lung) | GSM640581(GSM640581) small RNA in the liver with paternal control. (liver) | SRR346414(SRX098255) Global profiling of miRNA and the hairpin pre. (Spleen) | SRR037930(GSM510468) e7p5_rep4. (embryo) | SRR039186(GSM485326) shLuc.1309;ROSA-rtTA (+dox). (fibroblast) | GSM640580(GSM640580) small RNA in the liver with paternal Low pro. (liver) | Ago3IP812(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | SRR037927(GSM510465) e7p5_rep1. (embryo) | SRR346425(SRX098267) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR023851(GSM307160) ZHBT-c412hsmallrna_rep1. (cell line) | SRR059770(GSM562832) Treg_Dicer. (spleen) | SRR023850(GSM307159) ZHBT-c40hsmallrna_rep1. (cell line) | SRR042474(GSM539866) mouse embryonic stem cells [09-002]. (ESC) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR042475(GSM539867) mouse embryonic fibroblast cells [09-002]. (fibroblast) | SRR014229(GSM319953) 10 dpp MILI. (mili testes) | SRR306531(GSM750574) 19-24nt. (ago2 brain) | SRR014231(GSM319955) 16.5 dpc total. (testes) | SRR042471(GSM539863) mouse TFHS cells [09-002]. (spleen) | SRR346421(SRX098263) Global profiling of miRNA and the hairpin pre. (mixture) | SRR042477(GSM539869) mouse brain tissue [09-002]. (brain) | SRR042470(GSM539862) mouse Th17 cells [09-002]. (lymph) | SRR077864(GSM637801) 18-30 nt small RNAs. (liver) | GSM475281(GSM475281) total RNA. (testes) | GSM361398(GSM361398) CGNP_P6_p53--_Ink4c--_rep1. (brain) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR042481(GSM539873) mouse pancreatic tissue [09-002]. (pancreas) | SRR306527(GSM750570) 19-24nt. (ago2 brain) | SRR306543(GSM750586) 19-24nt. (ago2 brain) | GSM509279(GSM509279) MVH-/- E16.5 small RNA. (testes) | SRR306530(GSM750573) 19-24nt. (ago2 brain) | SRR038741(GSM527276) small RNA-Seq. (brain) | SRR346413(SRX098254) Global profiling of miRNA and the hairpin pre. (Heart) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .GATCTAGAAATAATTGTG....................................................................................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | |
| ....................................................................................................................TCAGAGGCATAAAGGTCATTTG................................................................................................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |