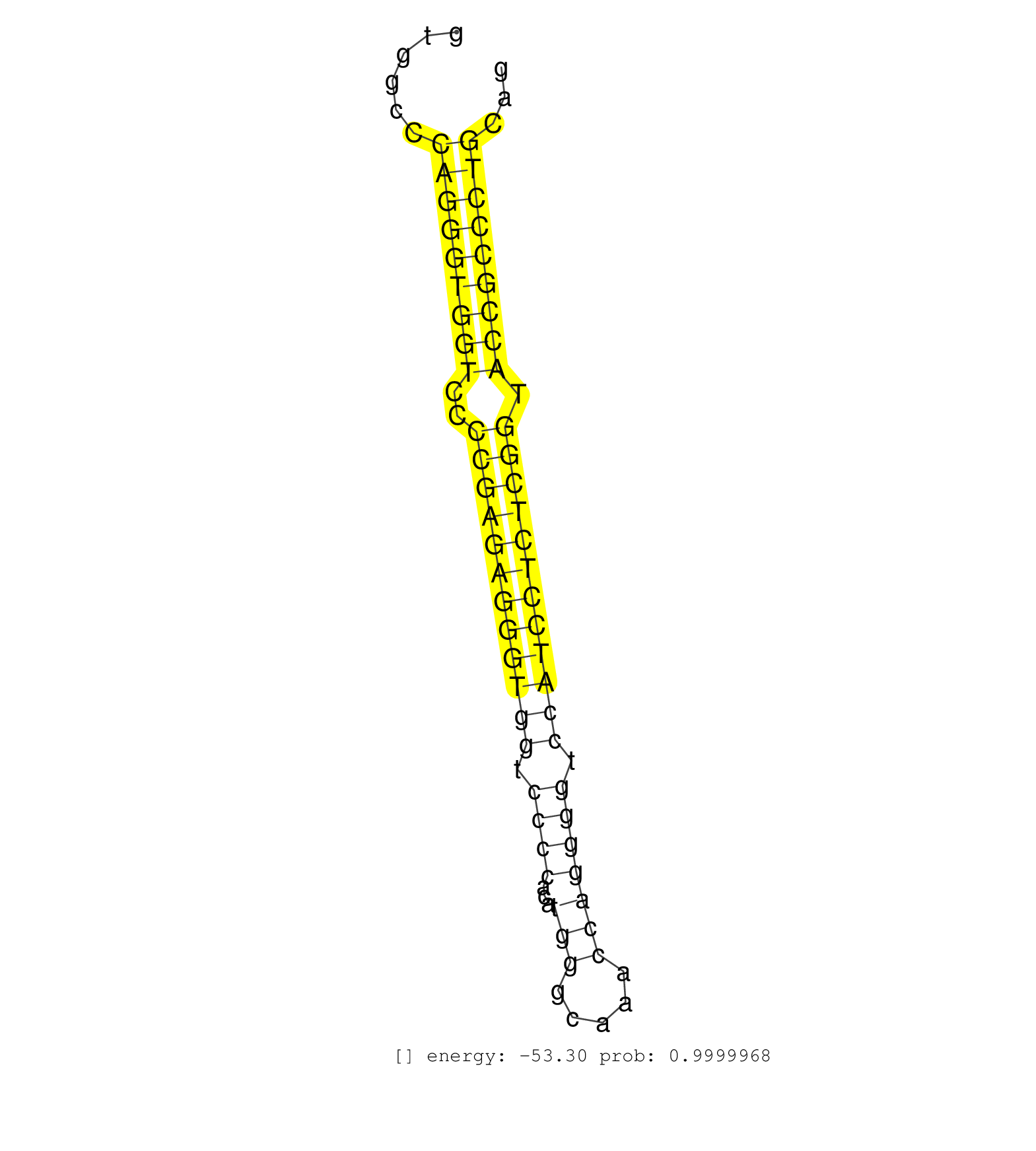
| (1) AGO.mut | (2) AGO1.ip | (16) AGO2.ip | (1) AGO3.ip | (11) B-CELL | (28) BRAIN | (3) CELL-LINE | (1) DCR.mut | (1) DGCR8.mut | (7) EMBRYO | (7) ESC | (4) FIBROBLAST | (1) HEART | (1) KIDNEY | (8) LIVER | (1) LUNG | (1) LYMPH | (10) OTHER | (5) OTHER.mut | (1) PANCREAS | (1) PIWI.mut | (4) SKIN | (9) SPLEEN | (10) TESTES | (2) THYMUS | (4) TOTAL-RNA | (1) UTERUS |
| AGAGAGAGGTGGAGACCCTCCGTGCTGAGCTGGAGAAGATTAAGATGGAGGTGAGGGGCGTGGCCCAGGGTGGTCCCCGAGAGGGTGGTCCCCACATGGGCAAACCAGGGGTCCATCCTCTCGGTACCGCCCTGCAGGCACAGCGGTACATCTCCCAGCTGAAGGGCCAGGTGAATGGCCTGGAGGC .................................................................(((((((((..((((((((((((.((((...(((.....))))))).)))))))))))).)))))))))..................................................... ...........................................................60...........................................................................137................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR042447(GSM539839) mouse mature B cells (spleen) replicate 2 [09-002]. (b cell) | SRR553583(SRX182789) source: Cerebellum. (Cerebellum) | SRR059769(GSM562831) Treg_Drosha. (spleen) | SRR042471(GSM539863) mouse TFHS cells [09-002]. (spleen) | SRR306542(GSM750585) 19-24nt. (ago2 brain) | mjLiverWT1() Liver Data. (liver) | SRR306540(GSM750583) 19-24nt. (ago2 brain) | SRR065059(SRR065059) Tissue-specific Regulation of Mouse MicroRNA . (pancreas) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (ago2 liver) | SRR042450(GSM539842) mouse B cells activated in vitro with LPS/IL4 replicate 3 [09-002]. (b cell) | SRR306531(GSM750574) 19-24nt. (ago2 brain) | SRR042459(GSM539851) mouse neutrophil cells replicate 1 [09-002]. (blood) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR553584(SRX182790) source: Heart. (Heart) | SRR346417(SRX098258) Global profiling of miRNA and the hairpin pre. (Brain) | SRR306535(GSM750578) 19-24nt. (ago2 brain) | SRR346420(SRX098261) Global profiling of miRNA and the hairpin pre. (Uterus) | SRR390299(GSM849857) cell line: NIH-3T3cell type: fibroblastip ant. (ago2 fibroblast) | SRR059772(GSM562834) CD4_Drosha. (spleen) | SRR279906(GSM689056) cell type: mouse embryonic stem cellcell line. (ESC) | SRR306534(GSM750577) 19-24nt. (ago2 brain) | SRR306539(GSM750582) 19-24nt. (ago2 brain) | SRR059768(GSM562830) Treg_control. (spleen) | SRR346421(SRX098263) Global profiling of miRNA and the hairpin pre. (mixture) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | Ago1IP812(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | SRR042452(GSM539844) mouse B1 B cells [09-002]. (b cell) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | GSM510452(GSM510452) newborn_rep8. (total RNA) | SRR042467(GSM539859) mouse T cells activated in vitro with Concanavalin A [09-002]. (spleen) | SRR039183(GSM485323) shLuc.1309 (-dox). (fibroblast) | SRR042451(GSM539843) mouse B cells activated in vitro with LPS/aIgD-Dextran [09-002]. (b cell) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) | SRR306532(GSM750575) 19-24nt. (ago2 brain) | SRR039185(GSM485325) shLuc.1309;ROSA-rtTA (-dox). (fibroblast) | SRR306529(GSM750572) 19-24nt. (ago2 brain) | mjLiverKO1() Liver Data. (Zcchc11 liver) | SRR038745(GSM527280) small RNA-Seq. (dgcr8 brain) | SRR306544(GSM750587) 19-24nt. (ago2 brain) | SRR042446(GSM539838) mouse mature B cells (spleen) replicate 1 [09-002]. (b cell) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | Ago1IP517(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | Ago3IP805(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | SRR042449(GSM539841) mouse B cells activated in vitro with LPS/IL4 replicate 2 [09-002]. (b cell) | SRR077866(GSM637803) 18-30 nt small RNAs. (liver) | SRR077864(GSM637801) 18-30 nt small RNAs. (liver) | SRR402760(SRX117943) source: ES E14 male cells. (ESC) | SRR042454(GSM539846) mouse germinal center B cells (lymph node) [09-002]. (b cell) | SRR059765(GSM562827) DN3_control. (thymus) | SRR306533(GSM750576) 19-24nt. (ago2 brain) | SRR059766(GSM562828) DN3_Drosha. (thymus) | GSM640581(GSM640581) small RNA in the liver with paternal control. (liver) | Ago2IP517(Rui) Quantitative functions of Argonaute proteins . (ago2 skin) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR116846(GSM678422) Small RNA deep sequencing from mouse epididym. (epididymis) | SRR553586(SRX182792) source: Testis. (testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | GSM361395(GSM361395) CGNP_P6_wt_rep2. (brain) | SRR042448(GSM539840) mouse B cells activated in vitro with LPS/IL4 replicate 1 [09-002]. (b cell) | SRR306538(GSM750581) 19-24nt. (ago2 brain) | SRR065055(SRR065055) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR346419(SRX098260) Global profiling of miRNA and the hairpin pre. (Lung) | SRR029123(GSM416611) NIH3T3. (cell line) | SRR042445(GSM539837) mouse immature B cells [09-002]. (b cell) | SRR346425(SRX098267) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR059770(GSM562832) Treg_Dicer. (spleen) | SRR038741(GSM527276) small RNA-Seq. (brain) | SRR039154(GSM471931) HL1_siSrf2_smallRNAseq. (muscle) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR042466(GSM539858) mouse CD8+ T cells (spleen) [09-002]. (spleen) | SRR553582(SRX182788) source: Brain. (Brain) | SRR042453(GSM539845) mouse marginal zone B cells (spleen) [09-002]. (b cell) | GSM314559(GSM314559) ESC dgcr8 (454). (ESC) | SRR346415(SRX098256) source: Testis. (Testes) | SRR345205(SRX097266) source: size fractionated RNA from mouse hipp. (brain) | SRR039186(GSM485326) shLuc.1309;ROSA-rtTA (+dox). (fibroblast) | GSM261959(GSM261959) oocytesmallRNA-19to30. (oocyte) | GSM640578(GSM640578) small RNA in the liver with paternal control. (liver) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR037906(GSM510442) brain_rep3. (brain) | SRR279903(GSM689053) cell type: mouse embryonic stem cellcell line. (ESC) | GSM519104(GSM519104) mouse_granulocyte_nuclei_smallRNAs. (cell line) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM510444(GSM510444) brain_rep5. (brain) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR042463(GSM539855) mouse natural killer cells [09-002]. (spleen) | SRR042483(GSM539875) mouse muscle tissue [09-002]. (muscle) | SRR059771(GSM562833) CD4_control. (spleen) | SRR037912(GSM510449) newborn_rep5. (total RNA) | SRR042468(GSM539860) mouse Th1 cells [09-002]. (lymph) | GSM314557(GSM314557) ESC dgcr8 (Illumina). (ESC) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR346416(SRX098257) Global profiling of miRNA and the hairpin pre. (Kidney) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR038740(GSM527275) small RNA-Seq. (dicer brain) | GSM361402(GSM361402) CGNP_P6_p53--_Ink4c--_rep5. (brain) | SRR306527(GSM750570) 19-24nt. (ago2 brain) | SRR346423(SRX098265) Global profiling of miRNA and the hairpin pre. (Brain) | GSM510456(GSM510456) newborn_rep12. (total RNA) | SRR402761(SRX117944) source: ES E14 male cells. (ESC) | SRR037914(GSM510451) newborn_rep7. (total RNA) | SRR042456(GSM539848) mouse hematopoetic progenitor cells [09-002]. (bone marrow) | SRR042458(GSM539850) mouse basophil cells [09-002]. (b cell) | SRR685339(GSM1079783) Small RNAs (15-50 nts in length) from immorta. (cell line) | SRR345196(SRX097257) source: size fractionated RNA from mouse hipp. (brain) | SRR345204(SRX097265) source: size fractionated RNA from mouse hipp. (brain) | SRR037929(GSM510467) e7p5_rep3. (embryo) | SRR345199(SRX097260) source: size fractionated RNA from mouse hipp. (brain) | SRR306536(GSM750579) 19-24nt. (ago2 brain) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................ATCCTCTCGGTACCGCCCTGCA................................................... | 22 | 1 | 37.00 | 37.00 | 1.00 | 4.00 | 4.00 | 1.00 | 3.00 | - | 1.00 | - | - | - | 2.00 | - | - | - | - | 2.00 | - | 4.00 | - | - | - | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..................................................................................................................ATCCTCTCGGTACCGCCCTGC.................................................... | 21 | 1 | 37.00 | 37.00 | - | 6.00 | 2.00 | - | 1.00 | - | 2.00 | - | 2.00 | - | 1.00 | - | - | 2.00 | 2.00 | 1.00 | - | - | - | - | 3.00 | 1.00 | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| ..................................................................................................................ATCCTCTCGGTACCGCCCTGCAT.................................................. | 23 | 1 | 16.00 | 37.00 | 5.00 | - | 1.00 | 2.00 | - | - | - | 1.00 | - | - | 2.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................ATCCTCTCGGTACCGCCC....................................................... | 18 | 1 | 12.00 | 12.00 | 6.00 | - | - | 2.00 | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................ATCCTCTCGGTACCGCCCTGCC................................................... | 22 | 1 | 10.00 | 37.00 | - | - | - | - | - | 6.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................ATCCTCTCGGTACCGCCCTGCAG.................................................. | 23 | 1 | 10.00 | 10.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...............................................................CCCAGGGTGGTCCCCGAGAGGGTGGT.................................................................................................. | 26 | 1 | 9.00 | 9.00 | - | - | 1.00 | - | - | - | - | 4.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................ATCCTCTCGGTACCGCCCTGCAA.................................................. | 23 | 1 | 8.00 | 37.00 | - | - | 1.00 | - | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................ATCCTCTCGGTACCGCCCT...................................................... | 19 | 1 | 7.00 | 7.00 | 1.00 | - | 1.00 | 3.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................CCAGGGTGGTCCCCGAGAGGGT..................................................................................................... | 22 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................GCTGAGCTGGAGAAGATT.................................................................................................................................................. | 18 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 |
| .................................................................CAGGGTGGTCCCCGAGAGGGTGG................................................................................................... | 23 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................CCCAGGGTGGTCCCCGAGAG........................................................................................................ | 20 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGGGGCGTGGCCCAGGG..................................................................................................................... | 20 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................ACATCTCCCAGCTGAAGGGCCAGGT............... | 25 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................CCCAGGGTGGTCCCCGAGAGGGTGG................................................................................................... | 25 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................ATCCTCTCGGTACCGCCCTG..................................................... | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................ATCCTCTCGGTACCGCCCTGT.................................................... | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................CCAGGGTGGTCCCCGAGAGGGTT.................................................................................................... | 23 | 1 | 2.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................CCCAGGGTGGTCCCCGAGAGGGTT.................................................................................................... | 24 | 1 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................ATCCTCTCGGTACCGCCCTGCATA................................................. | 24 | 1 | 2.00 | 37.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................ATCCTCTCGGTACCGCCCTGCCT.................................................. | 23 | 1 | 2.00 | 37.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGGGGCGTGGCCCAGGGTGGTCCCCGAGAGGGTGGTCCCCACATGGGCAAACCAGGGGTCCATCCTCTCGGTACCGCCCTGCT................................................... | 86 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................CCAGGGTGGTCCCCGAGAGGGTGT................................................................................................... | 24 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................GAAGATTAAGATGGAG......................................................................................................................................... | 16 | 3 | 1.67 | 1.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................TGCTGAGCTGGAGAAGATTAAG............................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................CATGGGCAAACCAGGGGTCCAT....................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................ATCCTCTCGGTACCGCCCTGTA................................................... | 22 | 1 | 1.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................CCAGGGTGGTCCCCGAGAGGG...................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................CCAGGGTGGTCCCCGAGAGGGTGGT.................................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................AGCTGGAGAAGATTAAGATGGAGGCAC..................................................................................................................................... | 27 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................CGTGCTGAGCTGGAGA....................................................................................................................................................... | 16 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................CAGGGTGGTCCCCGAGAGGGTGAT.................................................................................................. | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................CAGGGTGGTCCCCGAGAGGG...................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................GAAGATTAAGATGGAGGCACA.................................................................................................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................ATCCTCTCGGTACCGCCCTTCA................................................... | 22 | 1 | 1.00 | 7.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................TGCTGAGCTGGAGAAGATTAAGATGG........................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................ATCCTCTCGGTACCGCCCTGCAGTT................................................ | 25 | 1 | 1.00 | 10.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................GTGCTGAGCTGGAGAAGA.................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................CAGGGTGGTCCCCGAGAGGGTGGTT................................................................................................. | 25 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................TCCATCCTCTCGGTACCGCC........................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................ATCCTCTCGGTACCGCCCTGCAGT................................................. | 24 | 1 | 1.00 | 10.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .......................GCTGAGCTGGAGAAGATTAAGATGGAG......................................................................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................AGGGTGGTCCCCGAGAGGGT..................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................CCCAGGGTGGTCCCCGAGAGGGT..................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ....................CGTGCTGAGCTGGAGAAGCATG................................................................................................................................................. | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................TCCTCTCGGTACCGCCCTGCAG.................................................. | 22 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................TCCGTGCTGAGCTGGAGAAGATTAAGAT............................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................ATCCTCTCGGTACCGCCCTGCAAT................................................. | 24 | 1 | 1.00 | 37.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....AGAGGTGGAGACCCTCCGTGCTGAGCTGGAG........................................................................................................................................................ | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................CAGGGTGGTCCCCGAGAGGGTGGA.................................................................................................. | 24 | 1 | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................GTGCTGAGCTGGAGAAGATTA................................................................................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................ATCCTCTCGGTACCGCCCTGCAC.................................................. | 23 | 1 | 1.00 | 37.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................GGGTGGTCCCCGAGAGGGTGGT.................................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................CCAGGGTGGTCCCCGAGAGG....................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................AGCGGTACATCTCCCAGCTGAAGGGCCA.................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................ATCCTCTCGGTACCGCCCTGCATT................................................. | 24 | 1 | 1.00 | 37.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................GTGCTGAGCTGGAGAAGATA.................................................................................................................................................. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................GCTGAGCTGGAGAAGATTAAGATGGA.......................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................CCAGGGTGGTCCCCGAGAG........................................................................................................ | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................CGTGCTGAGCTGGAGAAGCA................................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | |
| ........................CTGAGCTGGAGAAGAATT................................................................................................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | |
| .......................GCTGAGCTGGAGAAGATTAAGAT............................................................................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................CAGGGTGGTCCCCGAGAGGGTGT................................................................................................... | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................GGTCCCCGAGAGGGTGGTCC................................................................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGGGGCGTGGCCCAGGGTGGTCCCCGAGAGGGTGGTCCCCACATGGGCAAACCAGGGGTCCATCCTCTCGGTACCGCCCTGCAG.................................................. | 87 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................AGGGTGGTCCCCGAGAGCGTG.................................................................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................CAGGGTGGTCCCCGAGAG........................................................................................................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................CCCAGGGTGGTCCCCGAGAGG....................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................CCCAGGGTGGTCCCCGAGAGGGTGT................................................................................................... | 25 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................TGCTGAGCTGGAGAAGATT.................................................................................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................CCCAGGGTGGTCCCCGAGAGA....................................................................................................... | 21 | 1 | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................CGTGCTGAGCTGGAGAAGCAC.................................................................................................................................................. | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .GAGAGAGGTGGAGACCCTCCGTGCTGAGCTG........................................................................................................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................GCTGAGCTGGAGAAGATTAAG............................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................GCAAACCAGGGGTCCC........................................................................ | 16 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................CCCAGGGTGGTCCCCGAGAGAAA..................................................................................................... | 23 | 1 | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................ATCCTCTCGGTACCGCCCTGTCT.................................................. | 23 | 1 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................TGCTGAGCTGGAGAAGAT................................................................................................................................................... | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................ACATGGGCAAACCAGGG............................................................................. | 17 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AGAGAGAGGTGGAGACCCTCCGTGCTGAGCTGGAGAAGATTAAGATGGAGGTGAGGGGCGTGGCCCAGGGTGGTCCCCGAGAGGGTGGTCCCCACATGGGCAAACCAGGGGTCCATCCTCTCGGTACCGCCCTGCAGGCACAGCGGTACATCTCCCAGCTGAAGGGCCAGGTGAATGGCCTGGAGGC .................................................................(((((((((..((((((((((((.((((...(((.....))))))).)))))))))))).)))))))))..................................................... ...........................................................60...........................................................................137................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR042447(GSM539839) mouse mature B cells (spleen) replicate 2 [09-002]. (b cell) | SRR553583(SRX182789) source: Cerebellum. (Cerebellum) | SRR059769(GSM562831) Treg_Drosha. (spleen) | SRR042471(GSM539863) mouse TFHS cells [09-002]. (spleen) | SRR306542(GSM750585) 19-24nt. (ago2 brain) | mjLiverWT1() Liver Data. (liver) | SRR306540(GSM750583) 19-24nt. (ago2 brain) | SRR065059(SRR065059) Tissue-specific Regulation of Mouse MicroRNA . (pancreas) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (ago2 liver) | SRR042450(GSM539842) mouse B cells activated in vitro with LPS/IL4 replicate 3 [09-002]. (b cell) | SRR306531(GSM750574) 19-24nt. (ago2 brain) | SRR042459(GSM539851) mouse neutrophil cells replicate 1 [09-002]. (blood) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR553584(SRX182790) source: Heart. (Heart) | SRR346417(SRX098258) Global profiling of miRNA and the hairpin pre. (Brain) | SRR306535(GSM750578) 19-24nt. (ago2 brain) | SRR346420(SRX098261) Global profiling of miRNA and the hairpin pre. (Uterus) | SRR390299(GSM849857) cell line: NIH-3T3cell type: fibroblastip ant. (ago2 fibroblast) | SRR059772(GSM562834) CD4_Drosha. (spleen) | SRR279906(GSM689056) cell type: mouse embryonic stem cellcell line. (ESC) | SRR306534(GSM750577) 19-24nt. (ago2 brain) | SRR306539(GSM750582) 19-24nt. (ago2 brain) | SRR059768(GSM562830) Treg_control. (spleen) | SRR346421(SRX098263) Global profiling of miRNA and the hairpin pre. (mixture) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | Ago1IP812(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | SRR042452(GSM539844) mouse B1 B cells [09-002]. (b cell) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | GSM510452(GSM510452) newborn_rep8. (total RNA) | SRR042467(GSM539859) mouse T cells activated in vitro with Concanavalin A [09-002]. (spleen) | SRR039183(GSM485323) shLuc.1309 (-dox). (fibroblast) | SRR042451(GSM539843) mouse B cells activated in vitro with LPS/aIgD-Dextran [09-002]. (b cell) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) | SRR306532(GSM750575) 19-24nt. (ago2 brain) | SRR039185(GSM485325) shLuc.1309;ROSA-rtTA (-dox). (fibroblast) | SRR306529(GSM750572) 19-24nt. (ago2 brain) | mjLiverKO1() Liver Data. (Zcchc11 liver) | SRR038745(GSM527280) small RNA-Seq. (dgcr8 brain) | SRR306544(GSM750587) 19-24nt. (ago2 brain) | SRR042446(GSM539838) mouse mature B cells (spleen) replicate 1 [09-002]. (b cell) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | Ago1IP517(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | Ago3IP805(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | SRR042449(GSM539841) mouse B cells activated in vitro with LPS/IL4 replicate 2 [09-002]. (b cell) | SRR077866(GSM637803) 18-30 nt small RNAs. (liver) | SRR077864(GSM637801) 18-30 nt small RNAs. (liver) | SRR402760(SRX117943) source: ES E14 male cells. (ESC) | SRR042454(GSM539846) mouse germinal center B cells (lymph node) [09-002]. (b cell) | SRR059765(GSM562827) DN3_control. (thymus) | SRR306533(GSM750576) 19-24nt. (ago2 brain) | SRR059766(GSM562828) DN3_Drosha. (thymus) | GSM640581(GSM640581) small RNA in the liver with paternal control. (liver) | Ago2IP517(Rui) Quantitative functions of Argonaute proteins . (ago2 skin) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR116846(GSM678422) Small RNA deep sequencing from mouse epididym. (epididymis) | SRR553586(SRX182792) source: Testis. (testes) | SRR051941(GSM545785) 18-32 nt total small RNAs (Mov10l-/-). (mov10L testes) | GSM361395(GSM361395) CGNP_P6_wt_rep2. (brain) | SRR042448(GSM539840) mouse B cells activated in vitro with LPS/IL4 replicate 1 [09-002]. (b cell) | SRR306538(GSM750581) 19-24nt. (ago2 brain) | SRR065055(SRR065055) Tissue-specific Regulation of Mouse MicroRNA . (jejunum) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR346419(SRX098260) Global profiling of miRNA and the hairpin pre. (Lung) | SRR029123(GSM416611) NIH3T3. (cell line) | SRR042445(GSM539837) mouse immature B cells [09-002]. (b cell) | SRR346425(SRX098267) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR028732(GSM400969) Mili-Tdrd1 KO associated. (mili testes) | SRR059770(GSM562832) Treg_Dicer. (spleen) | SRR038741(GSM527276) small RNA-Seq. (brain) | SRR039154(GSM471931) HL1_siSrf2_smallRNAseq. (muscle) | SRR391850(GSM852145) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR042466(GSM539858) mouse CD8+ T cells (spleen) [09-002]. (spleen) | SRR553582(SRX182788) source: Brain. (Brain) | SRR042453(GSM539845) mouse marginal zone B cells (spleen) [09-002]. (b cell) | GSM314559(GSM314559) ESC dgcr8 (454). (ESC) | SRR346415(SRX098256) source: Testis. (Testes) | SRR345205(SRX097266) source: size fractionated RNA from mouse hipp. (brain) | SRR039186(GSM485326) shLuc.1309;ROSA-rtTA (+dox). (fibroblast) | GSM261959(GSM261959) oocytesmallRNA-19to30. (oocyte) | GSM640578(GSM640578) small RNA in the liver with paternal control. (liver) | SRR029038(GSM433290) 25dpp_hetero_tdrd6-KO. (tdrd6 testes) | SRR037906(GSM510442) brain_rep3. (brain) | SRR279903(GSM689053) cell type: mouse embryonic stem cellcell line. (ESC) | GSM519104(GSM519104) mouse_granulocyte_nuclei_smallRNAs. (cell line) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | GSM510444(GSM510444) brain_rep5. (brain) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR042463(GSM539855) mouse natural killer cells [09-002]. (spleen) | SRR042483(GSM539875) mouse muscle tissue [09-002]. (muscle) | SRR059771(GSM562833) CD4_control. (spleen) | SRR037912(GSM510449) newborn_rep5. (total RNA) | SRR042468(GSM539860) mouse Th1 cells [09-002]. (lymph) | GSM314557(GSM314557) ESC dgcr8 (Illumina). (ESC) | SRR029037(GSM433289) 18dpp_homo_tdrd6-KO. (tdrd6 testes) | SRR346416(SRX098257) Global profiling of miRNA and the hairpin pre. (Kidney) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR038740(GSM527275) small RNA-Seq. (dicer brain) | GSM361402(GSM361402) CGNP_P6_p53--_Ink4c--_rep5. (brain) | SRR306527(GSM750570) 19-24nt. (ago2 brain) | SRR346423(SRX098265) Global profiling of miRNA and the hairpin pre. (Brain) | GSM510456(GSM510456) newborn_rep12. (total RNA) | SRR402761(SRX117944) source: ES E14 male cells. (ESC) | SRR037914(GSM510451) newborn_rep7. (total RNA) | SRR042456(GSM539848) mouse hematopoetic progenitor cells [09-002]. (bone marrow) | SRR042458(GSM539850) mouse basophil cells [09-002]. (b cell) | SRR685339(GSM1079783) Small RNAs (15-50 nts in length) from immorta. (cell line) | SRR345196(SRX097257) source: size fractionated RNA from mouse hipp. (brain) | SRR345204(SRX097265) source: size fractionated RNA from mouse hipp. (brain) | SRR037929(GSM510467) e7p5_rep3. (embryo) | SRR345199(SRX097260) source: size fractionated RNA from mouse hipp. (brain) | SRR306536(GSM750579) 19-24nt. (ago2 brain) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................ATCCTCTCGGTACCGCCCTGCA................................................... | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................TCCCCGAGAGGGTGGTCCCCA............................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................CCCCGAGAGGGTGGTCCCCAT............................................................................................ | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................CAGCGGTACATCTCCAAGA............................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |