| (1) AGO.mut | (2) AGO1.ip | (5) AGO2.ip | (2) AGO3.ip | (2) B-CELL | (17) BRAIN | (2) CELL-LINE | (2) DCR.mut | (1) DGCR8.mut | (4) EMBRYO | (1) ESC | (4) HEART | (1) KIDNEY | (11) LIVER | (2) LUNG | (12) OTHER | (3) OTHER.mut | (1) PANCREAS | (1) PIWI.mut | (6) SKIN | (3) SPLEEN | (8) TESTES | (1) TOTAL-RNA | (1) UTERUS |
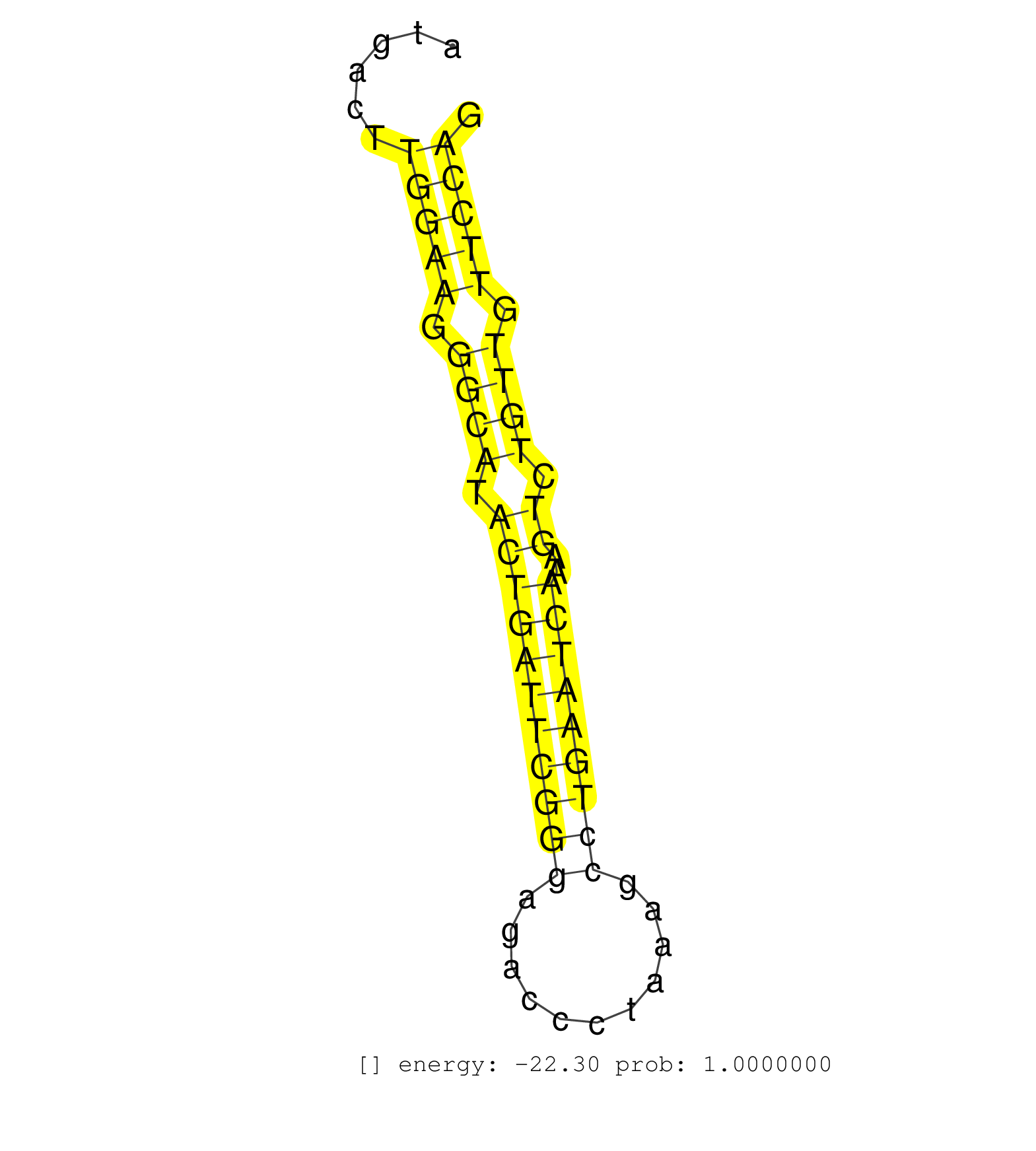
| TACCCTCAGCCCAGCGTGACATGGATGGAGTGCAGGGGCCACACCGATAGGTCAGTGGGCTTGGTTTTCCTCCTTAGGGTGAGTCTAAGTCTGGGTTGCGTACCCATGACTTGGAAGGGCATACTGATTCGGGAGACCCTAAAGCCTGAATCAAAGTCTGTTGTTCCAGGTGTGATGAAGCCCAGGCTTTGCAGGTTTGGAATGACACCCACCCTGAAG ...............................................................................................................(((((.((((.(((((((((((...........)))))))))..)).)))).)))))................................................... .........................................................................................................106............................................................169................................................ | Size | Perfect hit | Total Norm | Perfect Norm | mjLiverWT2() Liver Data. (liver) | mjLiverKO1() Liver Data. (Zcchc11 liver) | SRR346414(SRX098255) Global profiling of miRNA and the hairpin pre. (Spleen) | SRR042456(GSM539848) mouse hematopoetic progenitor cells [09-002]. (bone marrow) | SRR206942(GSM723283) other. (brain) | SRR038744(GSM527279) small RNA-Seq. (brain) | SRR206941(GSM723282) other. (brain) | SRR346418(SRX098259) Global profiling of miRNA and the hairpin pre. (Liver) | mjTestesWT1() Testes Data. (testes) | GSM510444(GSM510444) brain_rep5. (brain) | SRR206940(GSM723281) other. (brain) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | Ago2IP504(Rui) Quantitative functions of Argonaute proteins . (ago2 skin) | SRR553602(SRX182808) source: Sertoli cells. (Sertoli cells) | SRR346423(SRX098265) Global profiling of miRNA and the hairpin pre. (Brain) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR038743(GSM527278) small RNA-Seq. (dicer brain) | SRR042462(GSM539854) mouse macrophages [09-002]. (bone marrow) | Ago1IP517(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR073954(GSM629280) total RNA. (blood) | SRR095855BC7(SRX039178) sequencing of miRNA from wild type and diseas. (heart) | Ago3IP805(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | SRR073955(GSM629281) total RNA. (blood) | SRR042443(GSM539835) mouse pro B cells [09-002]. (b cell) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (ago2 liver) | SRR346419(SRX098260) Global profiling of miRNA and the hairpin pre. (Lung) | SRR039610(GSM527274) small RNA-Seq. (brain) | Ago2IP517(Rui) Quantitative functions of Argonaute proteins . (ago2 skin) | SRR553582(SRX182788) source: Brain. (Brain) | SRR095855BC2(SRX039178) sequencing of miRNA from wild type and diseas. (heart) | SRR042461(GSM539853) mouse dendritic cells [09-002]. (bone marrow) | Ago3IP812(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | SRR346417(SRX098258) Global profiling of miRNA and the hairpin pre. (Brain) | SRR306539(GSM750582) 19-24nt. (ago2 brain) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR042460(GSM539852) mouse neutrophil cells replicate 2 [09-002]. (blood) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR553585(SRX182791) source: Kidney. (Kidney) | SRR042459(GSM539851) mouse neutrophil cells replicate 1 [09-002]. (blood) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | SRR065047(SRR065047) Tissue-specific Regulation of Mouse MicroRNA . (liver) | GSM519104(GSM519104) mouse_granulocyte_nuclei_smallRNAs. (cell line) | SRR037921(GSM510459) e12p5_rep3. (embryo) | SRR042471(GSM539863) mouse TFHS cells [09-002]. (spleen) | GSM640579(GSM640579) small RNA in the liver with paternal Low pro. (liver) | GSM510456(GSM510456) newborn_rep12. (total RNA) | SRR077864(GSM637801) 18-30 nt small RNAs. (liver) | SRR037929(GSM510467) e7p5_rep3. (embryo) | SRR038740(GSM527275) small RNA-Seq. (dicer brain) | SRR042447(GSM539839) mouse mature B cells (spleen) replicate 2 [09-002]. (b cell) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR065058(SRR065058) Tissue-specific Regulation of Mouse MicroRNA . (pancreas) | SRR525243(SRA056111/SRX170319) Mus musculus domesticus miRNA sequencing. (ago2 Blood) | SRR042478(GSM539870) mouse lung tissue [09-002]. (lung) | SRR042479(GSM539871) mouse liver tissue [09-002]. (liver) | SRR306538(GSM750581) 19-24nt. (ago2 brain) | SRR039152(GSM471929) HL1_siNon_smallRNAseq. (muscle) | SRR023852(GSM307161) ZHBT-c424hsmallrna_rep1. (cell line) | SRR345207(SRX097268) source: size fractionated RNA from mouse hipp. (brain) | SRR032477(GSM485235) sRNA_activation_deep_sequencing. (uterus) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | Ago1IP812(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | SRR553584(SRX182790) source: Heart. (Heart) | SRR039154(GSM471931) HL1_siSrf2_smallRNAseq. (muscle) | SRR346415(SRX098256) source: Testis. (Testes) | SRR095853(SRX039174) sequencing of miRNA from wild type and diseas. (heart) | mjLiverWT3() Liver Data. (liver) | SRR059770(GSM562832) Treg_Dicer. (spleen) | SRR042484(GSM539876) mouse salivary gland tissue [09-002]. (salivary gland) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR037905(GSM510441) brain_rep2. (brain) | SRR042474(GSM539866) mouse embryonic stem cells [09-002]. (ESC) | SRR077866(GSM637803) 18-30 nt small RNAs. (liver) | mjTestesKO6() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................TGAATCAAAGTCTGTTGTTCCAG.................................................. | 23 | 1 | 85.00 | 85.00 | 30.00 | 13.00 | 4.00 | - | 5.00 | 2.00 | 3.00 | 4.00 | - | - | 5.00 | 2.00 | - | 3.00 | 1.00 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | 2.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................TGAATCAAAGTCTGTTGTTCCAGT................................................. | 24 | 1 | 20.00 | 85.00 | - | - | - | - | - | - | - | 1.00 | - | 2.00 | - | 1.00 | 2.00 | - | - | - | - | - | - | 1.00 | 1.00 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................TTGGAAGGGCATACTGATTCGG....................................................................................... | 22 | 1 | 15.00 | 15.00 | - | - | - | 1.00 | - | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | 1.00 | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - |
| ..............................................................................................................TTGGAAGGGCATACTGATTCGGG...................................................................................... | 23 | 1 | 14.00 | 14.00 | - | - | - | 3.00 | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | 2.00 | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..................................................................................................................................................TGAATCAAAGTCTGTTGTTC..................................................... | 20 | 1 | 8.00 | 8.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - |
| ..................................................................................................................................................TGAATCAAAGTCTGTTGTTCC.................................................... | 21 | 1 | 7.00 | 7.00 | - | - | 2.00 | - | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................TGACATGGATGGAGTGCAGGGGCCAC................................................................................................................................................................................. | 26 | 1 | 5.00 | 5.00 | - | - | - | - | - | - | - | - | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................TGAATCAAAGTCTGTTGTTCCA................................................... | 22 | 1 | 5.00 | 5.00 | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................TTGGAAGGGCATACTGATTCG........................................................................................ | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................TGAATCAAAGTCTGTTGTT...................................................... | 19 | 2 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - |
| ...................................................................................................................................................GAATCAAAGTCTGTTGTTCCAGT................................................. | 23 | 2.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................TGAATCAAAGTCTGTTGTTCCCG.................................................. | 23 | 1 | 2.00 | 7.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................TGAATCAAAGTCTGTTGTTCCAGA................................................. | 24 | 1 | 2.00 | 85.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..............................................................................................................TTGGAAGGGCATACTGATTCTGG...................................................................................... | 23 | 1 | 2.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................ACTTGGAAGGGCATACTGATTCG........................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................TTGGAAGGGCATACTGATTC......................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................TTGGAAGGGCATACTGATTCGGGAT.................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TGGAAGGGCATACTGATTCGG....................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................TTGGAAGGGCATACTGATTCGGGT..................................................................................... | 24 | 1 | 1.00 | 14.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................TTGGAAGGGCATACTGATTCGGGA..................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................TGAATCAAAGTCTGTTGTTCCAGAA................................................ | 25 | 1 | 1.00 | 85.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TGGAAGGGCATACTGATTCGGG...................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................CTGAATCAAAGTCTGTTGTTCCAG.................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................GATGGAGTGCAGGGGCCACAC............................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................CTGAATCAAAGTCTGTTGTTC..................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...................................................................................TCTAAGTCTGGGTTGCG....................................................................................................................... | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................CTTGGAAGGGCATACTGATTCGG....................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................TGTGATGAAGCCCAGAAGC.............................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................TTGGAAGGGCATACTGATTCGGA...................................................................................... | 23 | 1 | 1.00 | 15.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................ACTTGGAAGGGCATACTGATTCGG....................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................TGAATCAAAGTCTGTTGTTCCCGA................................................. | 24 | 1 | 1.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................CTTAGGGTGAGTCTATATC................................................................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................TGAATCAAAGTCTGTTG........................................................ | 17 | 2 | 0.50 | 0.50 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................GGGTGAGTCTAAGTCT............................................................................................................................... | 16 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 |
| TACCCTCAGCCCAGCGTGACATGGATGGAGTGCAGGGGCCACACCGATAGGTCAGTGGGCTTGGTTTTCCTCCTTAGGGTGAGTCTAAGTCTGGGTTGCGTACCCATGACTTGGAAGGGCATACTGATTCGGGAGACCCTAAAGCCTGAATCAAAGTCTGTTGTTCCAGGTGTGATGAAGCCCAGGCTTTGCAGGTTTGGAATGACACCCACCCTGAAG ...............................................................................................................(((((.((((.(((((((((((...........)))))))))..)).)))).)))))................................................... .........................................................................................................106............................................................169................................................ | Size | Perfect hit | Total Norm | Perfect Norm | mjLiverWT2() Liver Data. (liver) | mjLiverKO1() Liver Data. (Zcchc11 liver) | SRR346414(SRX098255) Global profiling of miRNA and the hairpin pre. (Spleen) | SRR042456(GSM539848) mouse hematopoetic progenitor cells [09-002]. (bone marrow) | SRR206942(GSM723283) other. (brain) | SRR038744(GSM527279) small RNA-Seq. (brain) | SRR206941(GSM723282) other. (brain) | SRR346418(SRX098259) Global profiling of miRNA and the hairpin pre. (Liver) | mjTestesWT1() Testes Data. (testes) | GSM510444(GSM510444) brain_rep5. (brain) | SRR206940(GSM723281) other. (brain) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | Ago2IP504(Rui) Quantitative functions of Argonaute proteins . (ago2 skin) | SRR553602(SRX182808) source: Sertoli cells. (Sertoli cells) | SRR346423(SRX098265) Global profiling of miRNA and the hairpin pre. (Brain) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR038743(GSM527278) small RNA-Seq. (dicer brain) | SRR042462(GSM539854) mouse macrophages [09-002]. (bone marrow) | Ago1IP517(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR073954(GSM629280) total RNA. (blood) | SRR095855BC7(SRX039178) sequencing of miRNA from wild type and diseas. (heart) | Ago3IP805(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | SRR073955(GSM629281) total RNA. (blood) | SRR042443(GSM539835) mouse pro B cells [09-002]. (b cell) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (ago2 liver) | SRR346419(SRX098260) Global profiling of miRNA and the hairpin pre. (Lung) | SRR039610(GSM527274) small RNA-Seq. (brain) | Ago2IP517(Rui) Quantitative functions of Argonaute proteins . (ago2 skin) | SRR553582(SRX182788) source: Brain. (Brain) | SRR095855BC2(SRX039178) sequencing of miRNA from wild type and diseas. (heart) | SRR042461(GSM539853) mouse dendritic cells [09-002]. (bone marrow) | Ago3IP812(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | SRR346417(SRX098258) Global profiling of miRNA and the hairpin pre. (Brain) | SRR306539(GSM750582) 19-24nt. (ago2 brain) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR042460(GSM539852) mouse neutrophil cells replicate 2 [09-002]. (blood) | mjTestesKO7() Testes Data. (Zcchc11 testes) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR553585(SRX182791) source: Kidney. (Kidney) | SRR042459(GSM539851) mouse neutrophil cells replicate 1 [09-002]. (blood) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | SRR065047(SRR065047) Tissue-specific Regulation of Mouse MicroRNA . (liver) | GSM519104(GSM519104) mouse_granulocyte_nuclei_smallRNAs. (cell line) | SRR037921(GSM510459) e12p5_rep3. (embryo) | SRR042471(GSM539863) mouse TFHS cells [09-002]. (spleen) | GSM640579(GSM640579) small RNA in the liver with paternal Low pro. (liver) | GSM510456(GSM510456) newborn_rep12. (total RNA) | SRR077864(GSM637801) 18-30 nt small RNAs. (liver) | SRR037929(GSM510467) e7p5_rep3. (embryo) | SRR038740(GSM527275) small RNA-Seq. (dicer brain) | SRR042447(GSM539839) mouse mature B cells (spleen) replicate 2 [09-002]. (b cell) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR065058(SRR065058) Tissue-specific Regulation of Mouse MicroRNA . (pancreas) | SRR525243(SRA056111/SRX170319) Mus musculus domesticus miRNA sequencing. (ago2 Blood) | SRR042478(GSM539870) mouse lung tissue [09-002]. (lung) | SRR042479(GSM539871) mouse liver tissue [09-002]. (liver) | SRR306538(GSM750581) 19-24nt. (ago2 brain) | SRR039152(GSM471929) HL1_siNon_smallRNAseq. (muscle) | SRR023852(GSM307161) ZHBT-c424hsmallrna_rep1. (cell line) | SRR345207(SRX097268) source: size fractionated RNA from mouse hipp. (brain) | SRR032477(GSM485235) sRNA_activation_deep_sequencing. (uterus) | SRR014230(GSM319954) 10 dpp Dnmt3L-KO MILI. (mili testes) | Ago1IP812(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | SRR553584(SRX182790) source: Heart. (Heart) | SRR039154(GSM471931) HL1_siSrf2_smallRNAseq. (muscle) | SRR346415(SRX098256) source: Testis. (Testes) | SRR095853(SRX039174) sequencing of miRNA from wild type and diseas. (heart) | mjLiverWT3() Liver Data. (liver) | SRR059770(GSM562832) Treg_Dicer. (spleen) | SRR042484(GSM539876) mouse salivary gland tissue [09-002]. (salivary gland) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR037905(GSM510441) brain_rep2. (brain) | SRR042474(GSM539866) mouse embryonic stem cells [09-002]. (ESC) | SRR077866(GSM637803) 18-30 nt small RNAs. (liver) | mjTestesKO6() Testes Data. (Zcchc11 testes) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................GGAGACCCTAAAGCCACC...................................................................... | 18 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................GGAGACCCTAAAGCCAAA...................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .ACCCTCAGCCCAGCGTG......................................................................................................................................................................................................... | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |