| (1) AGO.mut | (1) AGO1.ip | (20) AGO2.ip | (1) AGO3.ip | (42) BRAIN | (3) CELL-LINE | (2) DCR.mut | (1) DGCR8.mut | (9) EMBRYO | (14) ESC | (7) FIBROBLAST | (7) HEART | (5) LIVER | (1) LUNG | (1) LYMPH | (9) OTHER | (4) OTHER.mut | (3) SKIN | (1) SPLEEN | (11) TESTES | (2) TOTAL-RNA | (1) UTERUS |
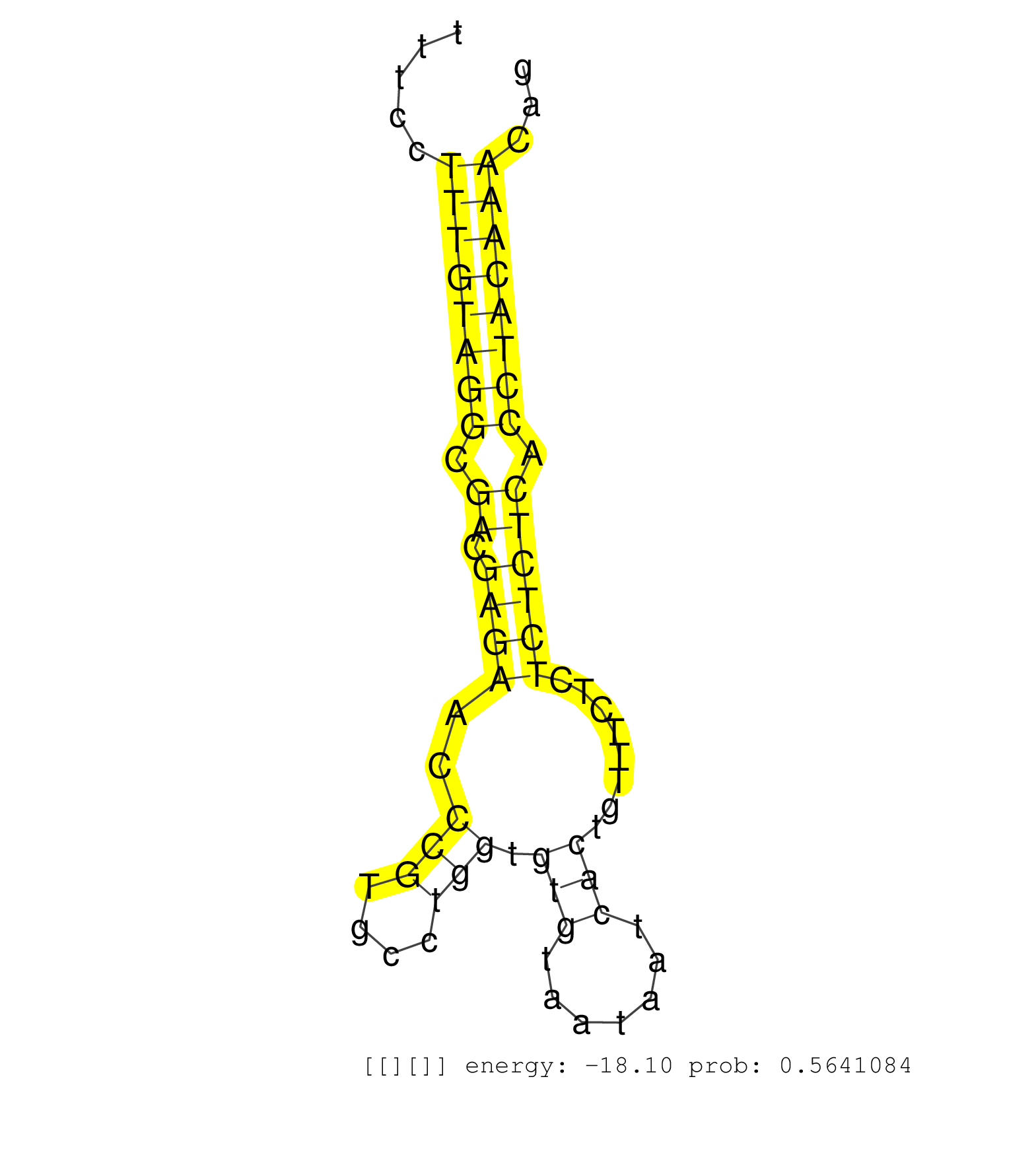
| CGGTATGTGCTCTAGAAGGGAGAGCCCCAGCAGTACCCAGTTTCGGCTTAGATGTGTTTGTCATCTAGAGTGTGGAAGGGAAGGCCGTAGCATCTGGTGACCTGTGTGTTCTTTGTGTCATTTAGCGTTTCCTTTGTAGGCGACGAGAACCCGTGCCTGGTGTGTAATAATCACTGTTTCTCTCTCTCACCTACAAACAGGTGAAGTTGGTGAACATCAGAAATGATGACATAACAGATGGAAATCCTAA ....................................................................................................................................((((((((.((.((((..(((....))).(((.......)))........)))))).))))))))..................................................... ...............................................................................................................................128.....................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR279907(GSM689057) cell type: mouse embryonic stem cellcell line. (ESC) | SRR546155(SRX180174) Global profiling of miRNA and the hairpin pre. (ago2 Neuroblastoma) | SRR346424(SRX098266) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR553582(SRX182788) source: Brain. (Brain) | SRR345203(SRX097264) source: size fractionated RNA from mouse hipp. (brain) | SRR345204(SRX097265) source: size fractionated RNA from mouse hipp. (brain) | SRR206942(GSM723283) other. (brain) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) | SRR391847(GSM852142) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR042474(GSM539866) mouse embryonic stem cells [09-002]. (ESC) | mjLiverKO2() Liver Data. (Zcchc11 liver) | SRR279903(GSM689053) cell type: mouse embryonic stem cellcell line. (ESC) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR346423(SRX098265) Global profiling of miRNA and the hairpin pre. (Brain) | SRR345202(SRX097263) source: size fractionated RNA from mouse hipp. (brain) | SRR206939(GSM723280) other. (brain) | SRR346417(SRX098258) Global profiling of miRNA and the hairpin pre. (Brain) | GSM314557(GSM314557) ESC dgcr8 (Illumina). (ESC) | SRR306534(GSM750577) 19-24nt. (ago2 brain) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR345196(SRX097257) source: size fractionated RNA from mouse hipp. (brain) | SRR345207(SRX097268) source: size fractionated RNA from mouse hipp. (brain) | SRR306533(GSM750576) 19-24nt. (ago2 brain) | SRR553583(SRX182789) source: Cerebellum. (Cerebellum) | SRR346425(SRX098267) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR306540(GSM750583) 19-24nt. (ago2 brain) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR306542(GSM750585) 19-24nt. (ago2 brain) | SRR206940(GSM723281) other. (brain) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | SRR394084(GSM855970) background strain: C57BL6/SV129cell type: KRa. (dicer cell line) | SRR402760(SRX117943) source: ES E14 male cells. (ESC) | SRR306535(GSM750578) 19-24nt. (ago2 brain) | SRR390299(GSM849857) cell line: NIH-3T3cell type: fibroblastip ant. (ago2 fibroblast) | SRR206941(GSM723282) other. (brain) | SRR345199(SRX097260) source: size fractionated RNA from mouse hipp. (brain) | SRR039185(GSM485325) shLuc.1309;ROSA-rtTA (-dox). (fibroblast) | SRR306532(GSM750575) 19-24nt. (ago2 brain) | SRR306537(GSM750580) 19-24nt. (ago2 brain) | SRR306531(GSM750574) 19-24nt. (ago2 brain) | GSM510444(GSM510444) brain_rep5. (brain) | SRR279906(GSM689056) cell type: mouse embryonic stem cellcell line. (ESC) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR345205(SRX097266) source: size fractionated RNA from mouse hipp. (brain) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR402761(SRX117944) source: ES E14 male cells. (ESC) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR279904(GSM689054) cell type: mouse embryonic stem cellcell line. (ESC) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) | SRR037906(GSM510442) brain_rep3. (brain) | mjTestesWT2() Testes Data. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR038744(GSM527279) small RNA-Seq. (brain) | SRR306536(GSM750579) 19-24nt. (ago2 brain) | SRR402766(SRX117949) source: ES PGK female cells. (ESC) | SRR345201(SRX097262) source: size fractionated RNA from mouse hipp. (brain) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR042475(GSM539867) mouse embryonic fibroblast cells [09-002]. (fibroblast) | SRR039184(GSM485324) shLuc.1309 (+dox). (fibroblast) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR038743(GSM527278) small RNA-Seq. (dicer brain) | SRR039183(GSM485323) shLuc.1309 (-dox). (fibroblast) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (ago2 liver) | SRR279905(GSM689055) cell type: mouse embryonic stem cellcell line. (ESC) | SRR042462(GSM539854) mouse macrophages [09-002]. (bone marrow) | SRR306539(GSM750582) 19-24nt. (ago2 brain) | SRR039154(GSM471931) HL1_siSrf2_smallRNAseq. (muscle) | GSM314552(GSM314552) ESC wild type (Illumina). (ESC) | SRR023851(GSM307160) ZHBT-c412hsmallrna_rep1. (cell line) | SRR306526(GSM750569) 19-24nt. (ago2 brain) | SRR095855BC1(SRX039178) sequencing of miRNA from wild type and diseas. (heart) | mjTestesWT1() Testes Data. (testes) | SRR390300(GSM849858) cell line: NIH-3T3cell type: fibroblastinfect. (ago2 fibroblast) | mjLiverKO1() Liver Data. (Zcchc11 liver) | SRR042471(GSM539863) mouse TFHS cells [09-002]. (spleen) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR553602(SRX182808) source: Sertoli cells. (Sertoli cells) | SRR042477(GSM539869) mouse brain tissue [09-002]. (brain) | SRR346419(SRX098260) Global profiling of miRNA and the hairpin pre. (Lung) | SRR346418(SRX098259) Global profiling of miRNA and the hairpin pre. (Liver) | SRR039610(GSM527274) small RNA-Seq. (brain) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR023847(GSM307156) mESv6.5smallrna_rep1. (cell line) | SRR095855BC5(SRX039178) sequencing of miRNA from wild type and diseas. (heart) | Ago1IP812(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | GSM314559(GSM314559) ESC dgcr8 (454). (ESC) | SRR095853(SRX039174) sequencing of miRNA from wild type and diseas. (heart) | GSM510453(GSM510453) newborn_rep9. (total RNA) | Ago3IP812(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | SRR039186(GSM485326) shLuc.1309;ROSA-rtTA (+dox). (fibroblast) | SRR095855BC2(SRX039178) sequencing of miRNA from wild type and diseas. (heart) | SRR042484(GSM539876) mouse salivary gland tissue [09-002]. (salivary gland) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR306529(GSM750572) 19-24nt. (ago2 brain) | SRR042468(GSM539860) mouse Th1 cells [09-002]. (lymph) | SRR039153(GSM471930) HL1_siSrf1_smallRNAseq. (muscle) | SRR095855BC8(SRX039178) sequencing of miRNA from wild type and diseas. (heart) | GSM509276(GSM509276) small RNA cloning by length. (testes) | Ago2IP504(Rui) Quantitative functions of Argonaute proteins . (ago2 skin) | SRR306544(GSM750587) 19-24nt. (ago2 brain) | SRR095855BC10(SRX039178) sequencing of miRNA from wild type and diseas. (heart) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR037914(GSM510451) newborn_rep7. (total RNA) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR037924(GSM510462) e9p5_rep2. (embryo) | SRR095855BC7(SRX039178) sequencing of miRNA from wild type and diseas. (heart) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR306527(GSM750570) 19-24nt. (ago2 brain) | SRR306543(GSM750586) 19-24nt. (ago2 brain) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR345206(SRX097267) source: size fractionated RNA from mouse hipp. (brain) | SRR346420(SRX098261) Global profiling of miRNA and the hairpin pre. (Uterus) | GSM314553(GSM314553) ESC dcr (Illumina). (ESC) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................TTTGTAGGCGACGAGAACCCGT................................................................................................ | 22 | 1 | 174.00 | 174.00 | 56.00 | 12.00 | 11.00 | 4.00 | 1.00 | 5.00 | 2.00 | 4.00 | - | 1.00 | - | - | - | 2.00 | 1.00 | 4.00 | 2.00 | 1.00 | 6.00 | - | - | 2.00 | 6.00 | 1.00 | 2.00 | 4.00 | - | 2.00 | 3.00 | - | 3.00 | - | 3.00 | - | 3.00 | 3.00 | 4.00 | - | - | 1.00 | 4.00 | 4.00 | 2.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - |
| ...................................................................................................................................CTTTGTAGGCGACGAGAACCCG................................................................................................. | 22 | 1 | 114.00 | 114.00 | 16.00 | 20.00 | 2.00 | 1.00 | 2.00 | 4.00 | 1.00 | 1.00 | 5.00 | - | - | 4.00 | - | - | - | 1.00 | 4.00 | 2.00 | - | 3.00 | - | 2.00 | - | 2.00 | 1.00 | - | 3.00 | 3.00 | - | 6.00 | 1.00 | 1.00 | - | - | - | - | 1.00 | 3.00 | - | 2.00 | - | - | - | 1.00 | - | 2.00 | 1.00 | 1.00 | - | 1.00 | 2.00 | - | - | 2.00 | - | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ....................................................................................................................................TTTGTAGGCGACGAGAACCCG................................................................................................. | 21 | 1 | 72.00 | 72.00 | 47.00 | 3.00 | 1.00 | 1.00 | 1.00 | - | 3.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TTTGTAGGCGACGAGAACCCGTG............................................................................................... | 23 | 1 | 49.00 | 49.00 | 20.00 | 4.00 | - | 3.00 | - | - | 3.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | 2.00 | - | - | - | 1.00 | 1.00 | - | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TTTGTAGGCGACGAGAACCCGTGT.............................................................................................. | 24 | 1 | 46.00 | 49.00 | 16.00 | 5.00 | 5.00 | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | 2.00 | - | - | - | 2.00 | - | 1.00 | 1.00 | - | - | 1.00 | - | - | 1.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................CTTTGTAGGCGACGAGAACCCGT................................................................................................ | 23 | 1 | 45.00 | 45.00 | 1.00 | 10.00 | 5.00 | 1.00 | 4.00 | 1.00 | - | - | 1.00 | - | - | - | - | 2.00 | - | - | 2.00 | - | - | - | - | - | - | 1.00 | 1.00 | 1.00 | 1.00 | - | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 2.00 | - | - | - | 1.00 | - | 1.00 | 1.00 | - | 2.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................CTTTGTAGGCGACGAGAACCCGA................................................................................................ | 23 | 1 | 43.00 | 114.00 | 4.00 | 5.00 | 2.00 | 2.00 | - | - | - | 1.00 | 3.00 | - | 12.00 | 1.00 | - | 1.00 | - | - | 1.00 | 1.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................CTTTGTAGGCGACGAGAACC................................................................................................... | 20 | 1 | 24.00 | 24.00 | 3.00 | 1.00 | 1.00 | - | 1.00 | - | 1.00 | - | - | 4.00 | - | 1.00 | 4.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TTTGTAGGCGACGAGAACCCGA................................................................................................ | 22 | 1 | 21.00 | 72.00 | 6.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................CTTTGTAGGCGACGAGAACCC.................................................................................................. | 21 | 1 | 20.00 | 20.00 | - | 5.00 | - | - | - | 2.00 | - | 3.00 | 2.00 | 1.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TTTGTAGGCGACGAGAACCCGTGA.............................................................................................. | 24 | 1 | 15.00 | 49.00 | 4.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TTGTAGGCGACGAGAACCCGTGT.............................................................................................. | 23 | 1 | 14.00 | 7.00 | 5.00 | - | 1.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - |
| ..................................................................................................................................CCTTTGTAGGCGACGAGAACCC.................................................................................................. | 22 | 1 | 14.00 | 14.00 | 10.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................TCCTTTGTAGGCGACGAGAACC................................................................................................... | 22 | 1 | 13.00 | 13.00 | 6.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................CTTTGTAGGCGACGAGAAC.................................................................................................... | 19 | 1 | 13.00 | 13.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | 6.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TTTGTAGGCGACGAGAACCC.................................................................................................. | 20 | 1 | 12.00 | 12.00 | 7.00 | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .....................................................................................................................................TTGTAGGCGACGAGAACCCGT................................................................................................ | 21 | 1 | 11.00 | 11.00 | 1.00 | - | - | 1.00 | - | 4.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................CCTTTGTAGGCGACGAGAACCCG................................................................................................. | 23 | 1 | 10.00 | 10.00 | - | 2.00 | - | - | 1.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................CCTTTGTAGGCGACGAGAACCCGT................................................................................................ | 24 | 1 | 8.00 | 8.00 | 1.00 | 3.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................CCTTTGTAGGCGACGAGAACC................................................................................................... | 21 | 1 | 8.00 | 8.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................CTTTGTAGGCGACGAGAA..................................................................................................... | 18 | 1 | 7.00 | 7.00 | - | 1.00 | - | - | - | - | - | - | - | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................................ATAACAGATGGAAATCCTAAGCTT | 24 | 7.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................TTGTAGGCGACGAGAACCCGTG............................................................................................... | 22 | 1 | 7.00 | 7.00 | 5.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................GGTGAACATCAGAAATGATGACAT.................. | 24 | 1 | 6.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................TCCTTTGTAGGCGACGAGAAC.................................................................................................... | 21 | 1 | 6.00 | 6.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................CCTTTGTAGGCGACGAGAAC.................................................................................................... | 20 | 1 | 5.00 | 5.00 | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TTTGTAGGCGACGAGAACCCGTA............................................................................................... | 23 | 1 | 5.00 | 174.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................ATGACATAACAGATGGA........ | 17 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................TCCTTTGTAGGCGACGAGAACCC.................................................................................................. | 23 | 1 | 4.00 | 4.00 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TTTGTAGGCGACGAGAACCCGTGC.............................................................................................. | 24 | 1 | 4.00 | 4.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................TGTAGGCGACGAGAACCCGTGT.............................................................................................. | 22 | 1 | 4.00 | 3.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................CTTTGTAGGCGACGAGAACCCGTT............................................................................................... | 24 | 1 | 4.00 | 45.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TTTGTAGGCGACGAGAAC.................................................................................................... | 18 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................AGGTGAAGTTGGTGAACA.................................. | 18 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................TCCTTTGTAGGCGACGAGA...................................................................................................... | 19 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TTTCTCTCTCTCACCTACAAAC.................................................... | 22 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TTTGTAGGCGACGAGAACCCGTGTA............................................................................................. | 25 | 1 | 3.00 | 49.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................CTTTGTAGGCGACGAGAACCCT................................................................................................. | 22 | 1 | 3.00 | 20.00 | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................TGTAGGCGACGAGAACCCGTG............................................................................................... | 21 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................CTTTGTAGGCGACGAGAACCCAT................................................................................................ | 23 | 1 | 3.00 | 20.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TTTGTAGGCGACGAGAACCCGTAA.............................................................................................. | 24 | 1 | 3.00 | 174.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................TGTAGGCGACGAGAACCCGT................................................................................................ | 20 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TTTCTCTCTCTCACCTACAAACAGT................................................. | 25 | 1 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TTCTCTCTCTCACCTACAAACAGA................................................. | 24 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................TTTGTAGGCGACGAGAACCCGAGT.............................................................................................. | 24 | 1 | 2.00 | 72.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................CTTTGTAGGCGACGAGAACCCGC................................................................................................ | 23 | 1 | 2.00 | 114.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................CTTTGTAGGCGACGAGAACCCGTGT.............................................................................................. | 25 | 2.00 | 0.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................TTTGTAGGCGACGAGAACCCGTT............................................................................................... | 23 | 1 | 2.00 | 174.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TTGTAGGCGACGAGAACCCGTGA.............................................................................................. | 23 | 1 | 2.00 | 7.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................TCCTTTGTAGGCGACGAGAA..................................................................................................... | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TTTGTAGGCGACGAGAACC................................................................................................... | 19 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TTGTAGGCGACGAGAACCCGTGAA............................................................................................. | 24 | 1 | 2.00 | 7.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................CTTTGTAGGCGACGAGAACCCTT................................................................................................ | 23 | 1 | 2.00 | 20.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................TCCTTTGTAGGCGACGAGAACCCGTGC.............................................................................................. | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................TTGGTGAACATCAGAAATGATGACA................... | 25 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................CTTTGTAGGCGACGAGAACCCGAT............................................................................................... | 24 | 1 | 2.00 | 114.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TTTGTAGGCGACGAGAACCCGAT............................................................................................... | 23 | 1 | 2.00 | 72.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................CTTTGTAGGCGACGAGAACCCCT................................................................................................ | 23 | 1 | 1.00 | 20.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................CCTTTGTAGGCGACGAGAACAA.................................................................................................. | 22 | 1 | 1.00 | 5.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TTCTCTCTCTCACCTACAAACAGT................................................. | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................CTTTGTAGGCGACGAGAACCCAA................................................................................................ | 23 | 1 | 1.00 | 20.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................TCTAGAGTGTGGAAGCGGT........................................................................................................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................CTTTGTAGGCGACGAGAACA................................................................................................... | 20 | 1 | 1.00 | 13.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................ATGATGACATAACAGATGGAAATCC... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TTTGTAGGCGACGAGAACCCGTGG.............................................................................................. | 24 | 1 | 1.00 | 49.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................CAGAAATGATGACATAACAG............. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................CTTTGTAGGCGACGAGAACCCGTTA.............................................................................................. | 25 | 1 | 1.00 | 45.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TTTGTAGGCGACGAGAACCCGTTTTT............................................................................................ | 26 | 1 | 1.00 | 174.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TTCTCTCTCTCACCTACAAACA................................................... | 22 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TTTGTAGGCGACGAGAACCCGTGTTTC........................................................................................... | 27 | 1 | 1.00 | 49.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................CTTTGTAGGCGACGAGAACCCGG................................................................................................ | 23 | 1 | 1.00 | 114.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................TCTAGAGTGTGGAAGCGGG........................................................................................................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................CTTTGTAGGCGACGAGAACCCA................................................................................................. | 22 | 1 | 1.00 | 20.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................................AATGATGACATAACAGATGGAAA...... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TTTCTCTCTCTCACCTACAAACA................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................CTTTGTAGGCGACGAGAACCA.................................................................................................. | 21 | 1 | 1.00 | 24.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................TACAAACAGGTGAAGTTCG........................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................................................................................GGTGAAGTTGGTGAATA.................................. | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................CTTTGTAGGCGACGAGAACCCGATT.............................................................................................. | 25 | 1 | 1.00 | 114.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................TGTAGGCGACGAGAACCCGTGC.............................................................................................. | 22 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................AGGTGAAGTTGGTGAACATC................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................TGAACATCAGAAATGATGAC.................... | 20 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................ATCTAGAGTGTGGAAGGG.......................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................CCTTTGTAGGCGACGAGAACCCGA................................................................................................ | 24 | 1 | 1.00 | 10.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TTGTAGGCGACGAGAACCCGTGTT............................................................................................. | 24 | 1 | 1.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................CCTTTGTAGGCGACGAGAACCCGTAA.............................................................................................. | 26 | 1 | 1.00 | 8.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TTTGTAGGCGACGAGAACCCCGT............................................................................................... | 23 | 1 | 1.00 | 12.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TTCTCTCTCTCACCTACAAAC.................................................... | 21 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TTTGTAGGCGACGAGAACCAGT................................................................................................ | 22 | 1 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TTTGTAGGCGACGAGAACCCGTAT.............................................................................................. | 24 | 1 | 1.00 | 174.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..................................................................................................................................CCTTTGTAGGCGACGAGAACA................................................................................................... | 21 | 1 | 1.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................TCTCTCACCTACAAATAG.................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................CTTTGTAGGCGACGAGAACCCGAAA.............................................................................................. | 25 | 1 | 1.00 | 114.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TTTGTAGGCGACGAGAACCCGTTT.............................................................................................. | 24 | 1 | 1.00 | 174.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................TACAAACAGGTGAAGTTGG........................................ | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................CCTTTGTAGGCGACGAGAACCAT................................................................................................. | 23 | 1 | 1.00 | 8.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................TACAAACAGGTGAAGTCGG........................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................................TTTCTCTCTCTCACCTACAAACAG.................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................CTTTGTAGGCGACGAGAACCCGATA.............................................................................................. | 25 | 1 | 1.00 | 114.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................................AGAAATGATGACATAACAGA............ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TTTGTAGGCGACGAGAACCCTTGA.............................................................................................. | 24 | 1 | 1.00 | 12.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................TACAAACAGGTGAAGTTGCGTA..................................... | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................................................................................................TGATGACATAACAGATGGACGCC.... | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................CTTTGTAGGCGACGAGAGC.................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................................................................GAAGTTGGTGAACATCAGAAA........................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................TCCTTTGTAGGCGACGT........................................................................................................ | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................TTTGTAGGCGACGAGAACCCT................................................................................................. | 21 | 1 | 1.00 | 12.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................GTTTCTCTCTCTCACCTACAAAC.................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................CCTTTGTAGGCGACGAGAATCC.................................................................................................. | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................CTTTGTAGGCGACGAGAACCAA................................................................................................. | 22 | 1 | 1.00 | 24.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................CCTTTGTAGGCGACGAGAAT.................................................................................................... | 20 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................TTTGTAGGCGACGAGAACCCGGGA.............................................................................................. | 24 | 1 | 1.00 | 72.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................GTTTCTCTCTCTCACCTACAAACA................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TTTGTAGGCGACGAGAACCCCT................................................................................................ | 22 | 1 | 1.00 | 12.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TTTGTAGGCGACGAGAACCCGCG............................................................................................... | 23 | 1 | 1.00 | 72.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................................................GAAGTTGGTGAACATCAGAA............................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TTTGTAGGCGACGAGAACCCTT................................................................................................ | 22 | 1 | 1.00 | 12.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................CTTTGTAGGCGACGAGAACCCGTTTA............................................................................................. | 26 | 1 | 1.00 | 45.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................................TAACAGATGGAAATCCT.. | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................................TGACATAACAGATGG......... | 15 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - |
| .......................................................................................................................................................................................................................ATCAGAAATGATGACAT.................. | 17 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................ACATCAGAAATGATGA..................... | 16 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 |
| CGGTATGTGCTCTAGAAGGGAGAGCCCCAGCAGTACCCAGTTTCGGCTTAGATGTGTTTGTCATCTAGAGTGTGGAAGGGAAGGCCGTAGCATCTGGTGACCTGTGTGTTCTTTGTGTCATTTAGCGTTTCCTTTGTAGGCGACGAGAACCCGTGCCTGGTGTGTAATAATCACTGTTTCTCTCTCTCACCTACAAACAGGTGAAGTTGGTGAACATCAGAAATGATGACATAACAGATGGAAATCCTAA ....................................................................................................................................((((((((.((.((((..(((....))).(((.......)))........)))))).))))))))..................................................... ...............................................................................................................................128.....................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR279907(GSM689057) cell type: mouse embryonic stem cellcell line. (ESC) | SRR546155(SRX180174) Global profiling of miRNA and the hairpin pre. (ago2 Neuroblastoma) | SRR346424(SRX098266) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR553582(SRX182788) source: Brain. (Brain) | SRR345203(SRX097264) source: size fractionated RNA from mouse hipp. (brain) | SRR345204(SRX097265) source: size fractionated RNA from mouse hipp. (brain) | SRR206942(GSM723283) other. (brain) | SRR402762(SRX117945) source: ES E14 male cells. (ESC) | SRR391847(GSM852142) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR042474(GSM539866) mouse embryonic stem cells [09-002]. (ESC) | mjLiverKO2() Liver Data. (Zcchc11 liver) | SRR279903(GSM689053) cell type: mouse embryonic stem cellcell line. (ESC) | SRR345198(SRX097259) source: size fractionated RNA from mouse hipp. (brain) | SRR346423(SRX098265) Global profiling of miRNA and the hairpin pre. (Brain) | SRR345202(SRX097263) source: size fractionated RNA from mouse hipp. (brain) | SRR206939(GSM723280) other. (brain) | SRR346417(SRX098258) Global profiling of miRNA and the hairpin pre. (Brain) | GSM314557(GSM314557) ESC dgcr8 (Illumina). (ESC) | SRR306534(GSM750577) 19-24nt. (ago2 brain) | SRR391851(GSM852146) gender: femalegenotype/variation: wild typeti. (embryo) | SRR345196(SRX097257) source: size fractionated RNA from mouse hipp. (brain) | SRR345207(SRX097268) source: size fractionated RNA from mouse hipp. (brain) | SRR306533(GSM750576) 19-24nt. (ago2 brain) | SRR553583(SRX182789) source: Cerebellum. (Cerebellum) | SRR346425(SRX098267) Global profiling of miRNA and the hairpin pre. (Neuroblastoma) | SRR306540(GSM750583) 19-24nt. (ago2 brain) | SRR391846(GSM852141) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR391845(GSM852140) gender: femalegenotype/variation: wild typeti. (embryo) | SRR248526(GSM733814) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR391852(GSM852147) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR306542(GSM750585) 19-24nt. (ago2 brain) | SRR206940(GSM723281) other. (brain) | SRR306541(GSM750584) 19-24nt. (ago2 brain) | SRR394084(GSM855970) background strain: C57BL6/SV129cell type: KRa. (dicer cell line) | SRR402760(SRX117943) source: ES E14 male cells. (ESC) | SRR306535(GSM750578) 19-24nt. (ago2 brain) | SRR390299(GSM849857) cell line: NIH-3T3cell type: fibroblastip ant. (ago2 fibroblast) | SRR206941(GSM723282) other. (brain) | SRR345199(SRX097260) source: size fractionated RNA from mouse hipp. (brain) | SRR039185(GSM485325) shLuc.1309;ROSA-rtTA (-dox). (fibroblast) | SRR306532(GSM750575) 19-24nt. (ago2 brain) | SRR306537(GSM750580) 19-24nt. (ago2 brain) | SRR306531(GSM750574) 19-24nt. (ago2 brain) | GSM510444(GSM510444) brain_rep5. (brain) | SRR279906(GSM689056) cell type: mouse embryonic stem cellcell line. (ESC) | SRR391848(GSM852143) gender: femalegenotype/variation: wild typeti. (embryo) | SRR345205(SRX097266) source: size fractionated RNA from mouse hipp. (brain) | SRR391853(GSM852148) gender: femalegenotype/variation: ADAR2-/-/AD. (embryo) | SRR402761(SRX117944) source: ES E14 male cells. (ESC) | SRR038742(GSM527277) small RNA-Seq. (dgcr8 brain) | SRR279904(GSM689054) cell type: mouse embryonic stem cellcell line. (ESC) | SRR345197(SRX097258) source: size fractionated RNA from mouse hipp. (brain) | SRR037906(GSM510442) brain_rep3. (brain) | mjTestesWT2() Testes Data. (testes) | SRR248525(GSM733813) cell type: Thy1- spermatogonial stem cellstra. (testes) | SRR038744(GSM527279) small RNA-Seq. (brain) | SRR306536(GSM750579) 19-24nt. (ago2 brain) | SRR402766(SRX117949) source: ES PGK female cells. (ESC) | SRR345201(SRX097262) source: size fractionated RNA from mouse hipp. (brain) | SRR345200(SRX097261) source: size fractionated RNA from mouse hipp. (brain) | SRR042475(GSM539867) mouse embryonic fibroblast cells [09-002]. (fibroblast) | SRR039184(GSM485324) shLuc.1309 (+dox). (fibroblast) | SRR029042(GSM433294) 18.5dpc_hetero_tdrd1-KO. (tdrd1 testes) | SRR038743(GSM527278) small RNA-Seq. (dicer brain) | SRR039183(GSM485323) shLuc.1309 (-dox). (fibroblast) | SRR040489(GSM533912) E18.5 Ago2 catalytically inactive LIVER small RNA. (ago2 liver) | SRR279905(GSM689055) cell type: mouse embryonic stem cellcell line. (ESC) | SRR042462(GSM539854) mouse macrophages [09-002]. (bone marrow) | SRR306539(GSM750582) 19-24nt. (ago2 brain) | SRR039154(GSM471931) HL1_siSrf2_smallRNAseq. (muscle) | GSM314552(GSM314552) ESC wild type (Illumina). (ESC) | SRR023851(GSM307160) ZHBT-c412hsmallrna_rep1. (cell line) | SRR306526(GSM750569) 19-24nt. (ago2 brain) | SRR095855BC1(SRX039178) sequencing of miRNA from wild type and diseas. (heart) | mjTestesWT1() Testes Data. (testes) | SRR390300(GSM849858) cell line: NIH-3T3cell type: fibroblastinfect. (ago2 fibroblast) | mjLiverKO1() Liver Data. (Zcchc11 liver) | SRR042471(GSM539863) mouse TFHS cells [09-002]. (spleen) | SRR248524(GSM733812) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR553602(SRX182808) source: Sertoli cells. (Sertoli cells) | SRR042477(GSM539869) mouse brain tissue [09-002]. (brain) | SRR346419(SRX098260) Global profiling of miRNA and the hairpin pre. (Lung) | SRR346418(SRX098259) Global profiling of miRNA and the hairpin pre. (Liver) | SRR039610(GSM527274) small RNA-Seq. (brain) | SRR391849(GSM852144) gender: femalegenotype/variation: ADAR2-/-tis. (embryo) | SRR023847(GSM307156) mESv6.5smallrna_rep1. (cell line) | SRR095855BC5(SRX039178) sequencing of miRNA from wild type and diseas. (heart) | Ago1IP812(Rui) Quantitative functions of Argonaute proteins . (ago1 skin) | GSM314559(GSM314559) ESC dgcr8 (454). (ESC) | SRR095853(SRX039174) sequencing of miRNA from wild type and diseas. (heart) | GSM510453(GSM510453) newborn_rep9. (total RNA) | Ago3IP812(Rui) Quantitative functions of Argonaute proteins . (ago3 skin) | SRR039186(GSM485326) shLuc.1309;ROSA-rtTA (+dox). (fibroblast) | SRR095855BC2(SRX039178) sequencing of miRNA from wild type and diseas. (heart) | SRR042484(GSM539876) mouse salivary gland tissue [09-002]. (salivary gland) | GSM509280(GSM509280) small RNA cloning by length. (testes) | SRR306529(GSM750572) 19-24nt. (ago2 brain) | SRR042468(GSM539860) mouse Th1 cells [09-002]. (lymph) | SRR039153(GSM471930) HL1_siSrf1_smallRNAseq. (muscle) | SRR095855BC8(SRX039178) sequencing of miRNA from wild type and diseas. (heart) | GSM509276(GSM509276) small RNA cloning by length. (testes) | Ago2IP504(Rui) Quantitative functions of Argonaute proteins . (ago2 skin) | SRR306544(GSM750587) 19-24nt. (ago2 brain) | SRR095855BC10(SRX039178) sequencing of miRNA from wild type and diseas. (heart) | SRR069809(GSM610965) small RNA sequencing; sample 1. (testes) | SRR037914(GSM510451) newborn_rep7. (total RNA) | SRR029041(GSM433293) 6w_homo_tdrd6-KO. (tdrd6 testes) | SRR037924(GSM510462) e9p5_rep2. (embryo) | SRR095855BC7(SRX039178) sequencing of miRNA from wild type and diseas. (heart) | SRR248523(GSM733811) cell type: Thy1+ spermatogonial stem cellstra. (testes) | SRR306527(GSM750570) 19-24nt. (ago2 brain) | SRR306543(GSM750586) 19-24nt. (ago2 brain) | SRR040488(GSM533911) E18.5 wild type LIVER small RNA. (liver) | SRR345206(SRX097267) source: size fractionated RNA from mouse hipp. (brain) | SRR346420(SRX098261) Global profiling of miRNA and the hairpin pre. (Uterus) | GSM314553(GSM314553) ESC dcr (Illumina). (ESC) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................GCGACGAGAACC................................................................................................... | 12 | 10 | 0.10 | 0.10 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.10 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |