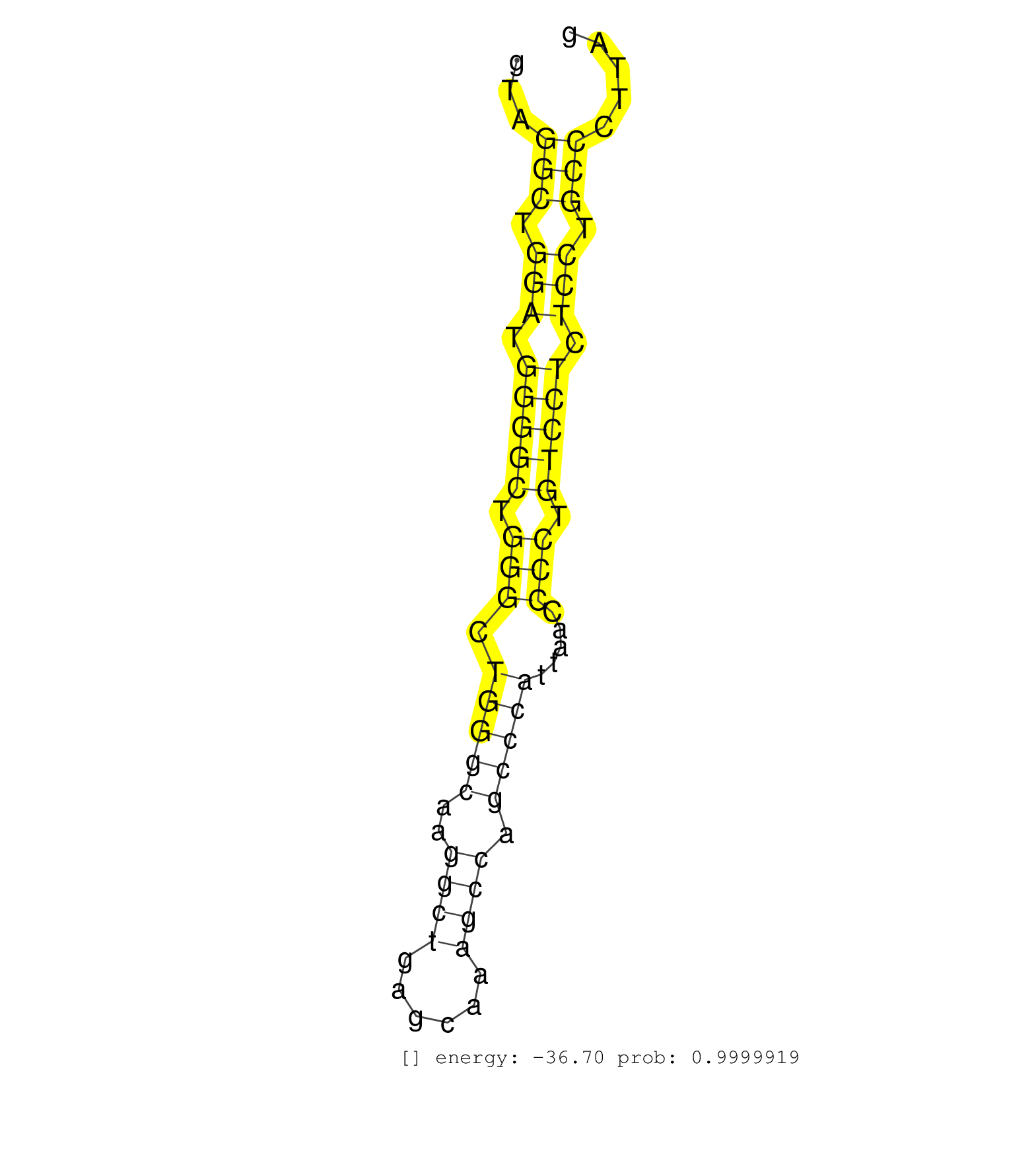
| (2) AGO2.ip | (4) B-CELL | (1) BRAIN | (2) BREAST | (8) CELL-LINE | (1) FIBROBLAST | (1) HELA | (1) LIVER | (1) OTHER |
| CCGGAGCTGCTCCCAGAGCAGCTGCTGCAAGATGCCTTCACTAGGCTCAGGTAGGCTGGATGGGGCTGGGCTGGGCAAGGCTGAGCAAAGCCAGCCCATTAACCCCTGTCCTCTCCTGCCCTTAGGGACATGCGGCTATCAATCACGGGGACCTTGGCAGAGAGCATTGTGGCTC .....................................................(((.(((.(((((.(((.(((((..((((......)))).))))).....))).))))).))).)))....................................................... ..................................................51........................................................................125................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | DRR001486(DRX001040) Hela long cytoplasmic cell fraction, LNA(+). (hela) | SRR387909(GSM843861) specific-host: Homo sapienshost cell line: Su. (cell line) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR037935(GSM510473) 293cand3. (cell line) | SRR033712(GSM497057) Burkitt Lymphoma (BL510). (B cell) | SRR060982(GSM569186) Human centrocyte [09-001]. (cell line) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | SRR189786 | SRR033715(GSM497060) Mantle Cell Lymphoma (Mino122). (B cell) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR037937(GSM510475) 293cand2. (cell line) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189787 | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR029127(GSM416756) A549. (cell line) | TAX577740(Rovira) total RNA. (breast) | SRR189784 | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | DRR000556(DRX000314) THP-1 whole cell RNA, after 3 day treatment w. (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................TAGGCTGGATGGGGCTGGGCTGT..................................................................................................... | 23 | 1 | 12.00 | 1.00 | 12.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGCTGGATGGGGCTGGGCTGGGCAAGGCTGAGCAAAGCCAGCCCATTAACCCCTGTCCTCTCCTG......................................................... | 68 | 1 | 9.00 | 9.00 | - | 9.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TAGGCTGGATGGGGCTGGGCT....................................................................................................... | 21 | 1 | 8.00 | 8.00 | 1.00 | - | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................CCCCTGTCCTCTCCTGCCCTTA................................................... | 22 | 1 | 7.00 | 7.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TAGGCTGGATGGGGCTGGGCTGG..................................................................................................... | 23 | 1 | 7.00 | 7.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TAGGCTGGATGGGGCTGGGCTAAA.................................................................................................... | 24 | 1 | 5.00 | 8.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................CCCCTGTCCTCTCCTGCCCTTT................................................... | 22 | 5.00 | 0.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................AGGCTGGATGGGGCTGGGCTGT..................................................................................................... | 22 | 3.00 | 0.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................TAGGCTGGATGGGGCTGGGCTTA..................................................................................................... | 23 | 1 | 3.00 | 8.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................CTGCTGCAAGATGCCTTCACTAGGCTCAG............................................................................................................................. | 29 | 1 | 2.00 | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................TCCTCTCCTGCCCTTCTA................................................. | 18 | 2.00 | 0.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................AGGCTGGATGGGGCTGGGCTGGTTAA................................................................................................. | 26 | 1 | 2.00 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TAGGCTGGATGGGGCTGGGCTA...................................................................................................... | 22 | 1 | 2.00 | 8.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGCTGGATGGGGCTGGGCTG...................................................................................................... | 23 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................CGGCTATCAATCACGGGGACC...................... | 21 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................CCCCTGTCCTCTCCTGCCCTAT................................................... | 22 | 2.00 | 0.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................................CCTTGGCAGAGAGCATTGT..... | 19 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................ATCACGGGGACCTTGGCAGAGAGCATT....... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TAGGCTGGATGGGGCTGGGCTG...................................................................................................... | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................TCAGGTAGGCTGGATAGCT.............................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................CCCCTGTCCTCTCCTGCCCTTAT.................................................. | 23 | 1 | 1.00 | 7.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................TAGGCTGGATGGGGCTGGGCTGTAA................................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................AGGCTGGATGGGGCTGGGCTGGAAAT................................................................................................. | 26 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................AGGCTGGATGGGGCTGGCG........................................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | |
| .......................GCTGCAAGATGCCTTCACTAGGCTCAG............................................................................................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................GGGACCTTGGCAGAGAGCAT........ | 20 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................CCCCTGTCCTCTCCTGCCCTTTT.................................................. | 23 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................TAGGCTGGATGGGGCTGGGCTGGT.................................................................................................... | 24 | 1 | 1.00 | 7.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................CTGGGCAAGGCTGAGAAC....................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | |
| ...................................................TAGGCTGGATGGGGCTGGGCAGG..................................................................................................... | 23 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................GGCTCAGGTAGGCTGGATCGGT.............................................................................................................. | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | |
| ....................................................AGGCTGGATGGGGCTGGGCTGG..................................................................................................... | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTAGGCTGGATGGGGCTGG.......................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................GGCTGGATGGGGCTGGGCTGGGCAAGG............................................................................................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................GGCTGGATGGGGCTGGGCTT...................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | |
| ....AGCTGCTCCCAGAGCAG.......................................................................................................................................................... | 17 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - |
| ...................................................TAGGCTGGATGGGGC............................................................................................................. | 15 | 6 | 0.33 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................GCTCAGGTAGGCTGG.................................................................................................................... | 15 | 8 | 0.12 | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.12 | - |
| ..............................................................GGGCTGGGCTGGGCAAGG............................................................................................... | 18 | 10 | 0.10 | 0.10 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.10 |
| CCGGAGCTGCTCCCAGAGCAGCTGCTGCAAGATGCCTTCACTAGGCTCAGGTAGGCTGGATGGGGCTGGGCTGGGCAAGGCTGAGCAAAGCCAGCCCATTAACCCCTGTCCTCTCCTGCCCTTAGGGACATGCGGCTATCAATCACGGGGACCTTGGCAGAGAGCATTGTGGCTC .....................................................(((.(((.(((((.(((.(((((..((((......)))).))))).....))).))))).))).)))....................................................... ..................................................51........................................................................125................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | DRR001486(DRX001040) Hela long cytoplasmic cell fraction, LNA(+). (hela) | SRR387909(GSM843861) specific-host: Homo sapienshost cell line: Su. (cell line) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR037935(GSM510473) 293cand3. (cell line) | SRR033712(GSM497057) Burkitt Lymphoma (BL510). (B cell) | SRR060982(GSM569186) Human centrocyte [09-001]. (cell line) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | SRR189786 | SRR033715(GSM497060) Mantle Cell Lymphoma (Mino122). (B cell) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR037937(GSM510475) 293cand2. (cell line) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189787 | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR029127(GSM416756) A549. (cell line) | TAX577740(Rovira) total RNA. (breast) | SRR189784 | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | DRR000556(DRX000314) THP-1 whole cell RNA, after 3 day treatment w. (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................CATTAACCCCTGTCCTCA............................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................TCCTCTCCTGCCCTTAATA................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | |
| ...............................................................................GCTGAGCAAAGCCAGCCCG............................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | |
| .........................................................................................................................TTAGGGACATGCGGCTATGC.................................. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |