| (1) AGO1.ip | (2) AGO2.ip | (5) B-CELL | (2) BRAIN | (12) BREAST | (28) CELL-LINE | (2) CERVIX | (2) FIBROBLAST | (4) HEART | (3) HELA | (5) LIVER | (1) OTHER | (1) RRP40.ip | (26) SKIN | (1) UTERUS | (1) XRN.ip |
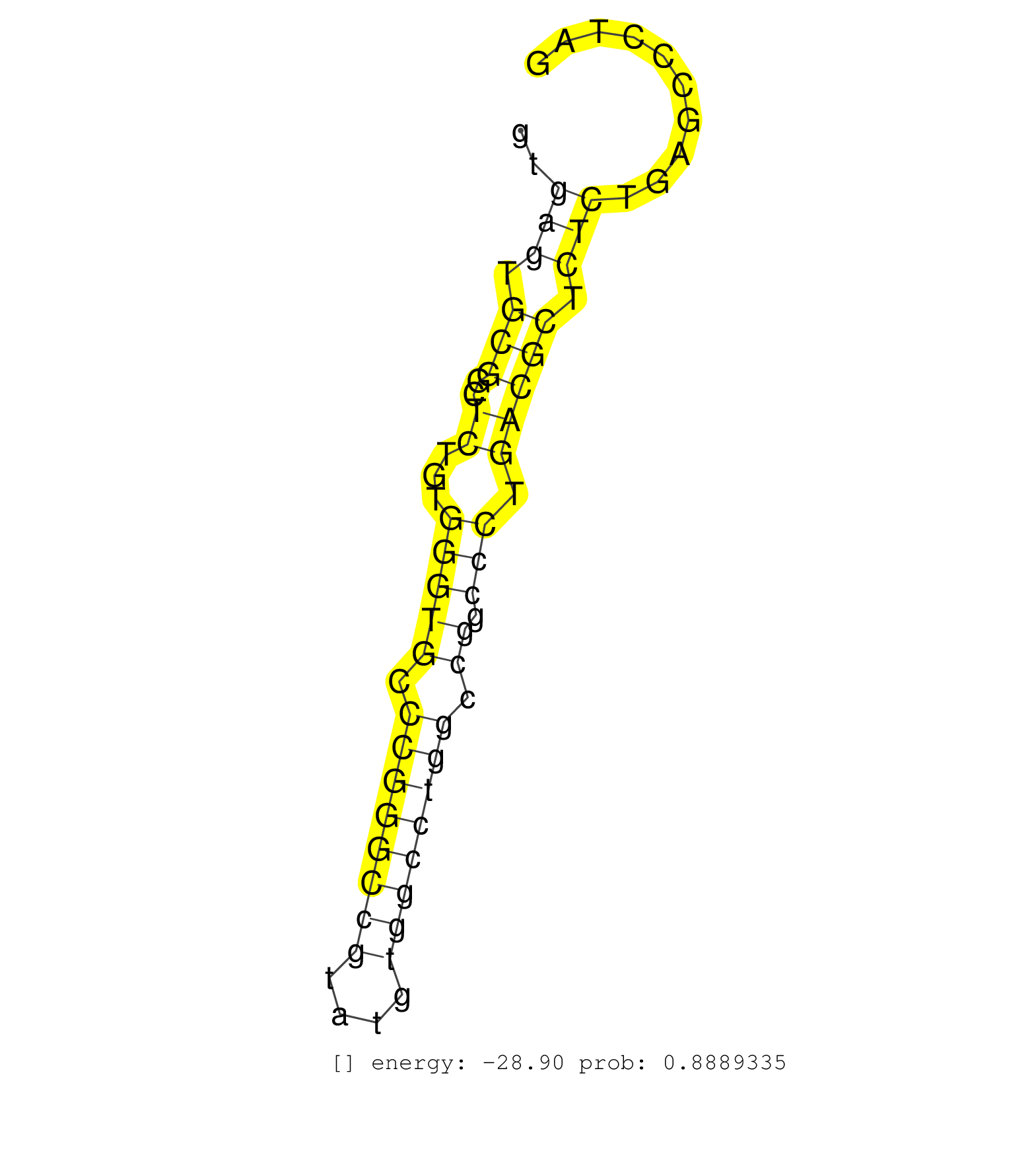
| GCTGGCGTGCGTTGCAACTCCATTCCGGCCGAGTACATCAGTGCTGAGAGGTGAGTGCGGCTCTGTGGGTGCCCGGGCCGTATGTGGCCTGGCCGGCCCTGACGCTCTCTGAGCCCTAGGATCGTGTGTGAGATGGAGGAGTCGCTGGTGCCCAGCCCGCCGCCGGGGC ....................................................(((.(((..((...(((((.((((((((....)))))))).)).))).))))).)))............................................................ ..................................................51..................................................................119................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | SRR037941(GSM510479) 293DroshaTN. (cell line) | SRR330871(SRX091709) tissue: skin psoriatic involveddisease state:. (skin) | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR326281(GSM769511) Dicer mRNA was knocked down using siDicer, cy. (cell line) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR033731(GSM497076) h929 Cell line (h929). (B cell) | SRR330897(SRX091735) tissue: skin psoriatic uninvolveddisease stat. (skin) | DRR001489(DRX001043) Hela short nuclear cell fraction, control. (hela) | SRR330923(SRX091761) tissue: normal skindisease state: normal. (skin) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR040035(GSM532920) G001T. (cervix) | SRR207110(GSM721072) Nuclear RNA. (cell line) | TAX577739(Rovira) total RNA. (breast) | SRR037937(GSM510475) 293cand2. (cell line) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR330913(SRX091751) tissue: normal skindisease state: normal. (skin) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577745(Rovira) total RNA. (breast) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) | SRR330882(SRX091720) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR326282(GSM769512) Dicer mRNA was knocked down using siDicer, to. (cell line) | SRR363673(GSM830250) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | DRR000558(DRX000316) THP-1 whole cell RNA, after 3 day treatment w. (cell line) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577744(Rovira) total RNA. (breast) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR330914(SRX091752) tissue: normal skindisease state: normal. (skin) | SRR330858(SRX091696) tissue: skin psoriatic involveddisease state:. (skin) | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | SRR330859(SRX091697) tissue: skin psoriatic involveddisease state:. (skin) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR191547(GSM715657) 155genomic small RNA (size selected RNA from . (breast) | TAX577453(Rovira) total RNA. (breast) | SRR191488(GSM715598) 14genomic small RNA (size selected RNA from t. (breast) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR037939(GSM510477) 293cand5_rep1. (cell line) | DRR000555(DRX000313) THP-1 whole cell RNA, no treatment. (cell line) | SRR189785 | SRR191622(GSM715732) 175genomic small RNA (size selected RNA from . (breast) | SRR191613(GSM715723) 66genomic small RNA (size selected RNA from t. (breast) | GSM1105753INPUT(GSM1105753) small RNA sequencing data. (hela) | SRR040039(GSM532924) G531T. (cervix) | TAX577741(Rovira) total RNA. (breast) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR038856(GSM458539) D11. (cell line) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR060983(GSM569187) Human pre-germinal center B cell [09-001]. (cell line) | SRR038857(GSM458540) D20. (cell line) | SRR330880(SRX091718) tissue: skin psoriatic involveddisease state:. (skin) | SRR330903(SRX091741) tissue: skin psoriatic uninvolveddisease stat. (skin) | DRR000559(DRX000317) THP-1 whole cell RNA, no treatment. (cell line) | GSM450598(GSM450598) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330885(SRX091723) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR033711(GSM497056) GCB DLBCL (GCB110). (B cell) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR060984(GSM569188) Human plasma cell [09-001]. (cell line) | SRR330916(SRX091754) tissue: normal skindisease state: normal. (skin) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | SRR038861(GSM458544) MM466. (cell line) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR033717(GSM497062) Mentle Cell Lymphoma (MCL112). (B cell) | SRR191565(GSM715675) 92genomic small RNA (size selected RNA from t. (breast) | SRR189786 | SRR191434(GSM715544) 171genomic small RNA (size selected RNA from . (breast) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | DRR000557(DRX000315) THP-1 whole cell RNA, after 3 day treatment w. (cell line) | SRR444049(SRX128897) Sample 9cDNABarcode: AF-PP-334: ACG CTC TTC C. (skin) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR330899(SRX091737) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR189784 | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | DRR001488(DRX001042) Hela short nuclear cell fraction, LNA(+). (hela) | SRR095854(SRX039177) miRNA were isolated from FirstChoice Human Br. (brain) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................TGCGGCTCTGTGGGTGCCCGGGC........................................................................................... | 23 | 1 | 18.00 | 18.00 | - | - | 2.00 | 1.00 | 3.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | 1.00 | 1.00 | - | - | - | - | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................TGCGGCTCTGTGGGTGCCCGGG............................................................................................ | 22 | 1 | 8.00 | 8.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................GCGGCTCTGTGGGTGCCCGGGC........................................................................................... | 22 | 1 | 8.00 | 8.00 | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................CGTATGTGGCCTGGCCGGC........................................................................ | 19 | 1 | 5.00 | 5.00 | - | - | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................CTGACGCTCTCTGAGCCCTAGT................................................. | 22 | 1 | 5.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................TGCGGCTCTGTGGGTGCCCGG............................................................................................. | 21 | 1 | 4.00 | 4.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................GTACATCAGTGCTGAGAG....................................................................................................................... | 18 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................CCGTATGTGGCCTGGCCGGCC....................................................................... | 21 | 1 | 3.00 | 3.00 | - | - | - | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................CTGACGCTCTCTGAGCCCTAGTT................................................ | 23 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TAGGATCGTGTGTGATACG.................................. | 19 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................TGCGGCTCTGTGGGTGCCCGGGA........................................................................................... | 23 | 1 | 2.00 | 8.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................GCGGCTCTGTGGGTGCCCGGG............................................................................................ | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................CTGACGCTCTCTGAGCCCTAG.................................................. | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................GCGGCTCTGTGGGTGCCCGGGCT.......................................................................................... | 23 | 1 | 2.00 | 8.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................GTGCTGAGAGGTGAGTGCGGCT........................................................................................................... | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................CCTGACGCTCTCTGAGCCCTAG.................................................. | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................TACATCAGTGCTGAGAG....................................................................................................................... | 17 | 2 | 1.50 | 1.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - |
| ...................................CATCAGTGCTGAGAGGTT.................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................TGCGGCTCTGTGGGTGCCCGGGG........................................................................................... | 23 | 1 | 1.00 | 8.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TCTGAGCCCTAGGATCCA............................................ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................GTGCGGCTCTGTGGGTGCCCGGGC........................................................................................... | 24 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TAGGATCGTGTGTGATACA.................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................TGCGGCTCTGTGGGTGCCCGGGTAAT........................................................................................ | 26 | 1 | 1.00 | 8.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................CGTATGTGGCCTGGCCGG......................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TAGGATCGTGTGTGATAG................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................CTGAGCCCTAGGATCACGT.......................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................GTGCGGCTCTGTGGGTGCCCGGG............................................................................................ | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................TGCGGCTCTGTGGGTGCCCGGT............................................................................................ | 22 | 1 | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................TCTGAGCCCTAGGATTGTG........................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................GATGGAGGAGTCGCTGGTGCCCAGCCCGC......... | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................TGCGGCTCTGTGGGTGCCCGGGCAT......................................................................................... | 25 | 1 | 1.00 | 18.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................CGGCTCTGTGGGTGCCCGGGCCTT........................................................................................ | 24 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................TGCGGCTCTGTGGGTGCCCGGGCTTAT....................................................................................... | 27 | 1 | 1.00 | 18.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................GCGGCTCTGTGGGTGCCCGGGG........................................................................................... | 22 | 1 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................CCCTGACGCTCTCTGAGCCCTCGT................................................. | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................TGGCCGGCCCTGACGCTCTCTGA......................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........CGTTGCAACTCCATTCCGGCCGAGTACA.................................................................................................................................... | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................TAGGATCGTGTGTGATAGG.................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................TGCGGCTCTGTGGGTGCCCGGGT........................................................................................... | 23 | 1 | 1.00 | 8.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................GTGCCCAGCCCGCCGAGC.... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................CCCTGACGCTCTCTGAGCCCTA................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................CGGCTCTGTGGGTGCCCGGGCC.......................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................CTCTGAGCCCTAGGATTGCA........................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................TGCGGCTCTGTGGGTGCAT............................................................................................... | 19 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - |
| ..................................................................................................CTGACGCTCTCTGAGCCCTAGA................................................. | 22 | 1 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................GCGGCTCTGTGGGTGCCCGGGTAT......................................................................................... | 24 | 1 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................TGCGGCTCTGTGGGTGCCCGGGAATT........................................................................................ | 26 | 1 | 1.00 | 8.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................GTGCGGCTCTGTGGGTGCCCGGGCC.......................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................CGGCTCTGTGGGTGCCCGGGCCAA........................................................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................CCCAGCCCGCCGCCGGGCG | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................AGTGCGGCTCTGTGGGTGCCCGGT............................................................................................ | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................CTGACGCTCTCTGAGCCCTAGTA................................................ | 23 | 1 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................GTGCGGCTCTGTGGGTGCCCGGT............................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................GCCCGCCGCCGGGGCG | 16 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................TGCGGCTCTGTGGGTGCCCGGGCAAT........................................................................................ | 26 | 1 | 1.00 | 18.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................CTGACGCTCTCTGAGCCCTATA................................................. | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................GTGCGGCTCTGTGGGTGCCCGG............................................................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................GCGGCTCTGTGGGTGCCCGGGCAA......................................................................................... | 24 | 1 | 1.00 | 8.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................CCCTGACGCTCTCTGAGCC...................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................AGTGCGGCTCTGTGGGTGCCCGGGCC.......................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................TGCGGCTCTGTGGGTGCCCGGAAGA......................................................................................... | 25 | 1 | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................GCCCGCCGCCGGGGCGCG | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................TGCGGCTCTGTGGGTGC................................................................................................. | 17 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................AGTGCGGCTCTGTGGGTGC................................................................................................. | 19 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................TGCGGCTCTGTGGGTGCAA............................................................................................... | 19 | 2 | 0.50 | 1.00 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................CGGCTCTGTGGGTGCC................................................................................................ | 16 | 2 | 0.50 | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................CATTCCGGCCGAG........................................................................................................................................ | 13 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - |
| .............................................................................................................................GTGTGAGATGGAGGAG............................ | 16 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................TCCGGCCGAGTAC..................................................................................................................................... | 13 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - | - | - | - |
| GCTGGCGTGCGTTGCAACTCCATTCCGGCCGAGTACATCAGTGCTGAGAGGTGAGTGCGGCTCTGTGGGTGCCCGGGCCGTATGTGGCCTGGCCGGCCCTGACGCTCTCTGAGCCCTAGGATCGTGTGTGAGATGGAGGAGTCGCTGGTGCCCAGCCCGCCGCCGGGGC ....................................................(((.(((..((...(((((.((((((((....)))))))).)).))).))))).)))............................................................ ..................................................51..................................................................119................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | SRR037941(GSM510479) 293DroshaTN. (cell line) | SRR330871(SRX091709) tissue: skin psoriatic involveddisease state:. (skin) | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR326281(GSM769511) Dicer mRNA was knocked down using siDicer, cy. (cell line) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR033731(GSM497076) h929 Cell line (h929). (B cell) | SRR330897(SRX091735) tissue: skin psoriatic uninvolveddisease stat. (skin) | DRR001489(DRX001043) Hela short nuclear cell fraction, control. (hela) | SRR330923(SRX091761) tissue: normal skindisease state: normal. (skin) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR040035(GSM532920) G001T. (cervix) | SRR207110(GSM721072) Nuclear RNA. (cell line) | TAX577739(Rovira) total RNA. (breast) | SRR037937(GSM510475) 293cand2. (cell line) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR330913(SRX091751) tissue: normal skindisease state: normal. (skin) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577745(Rovira) total RNA. (breast) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) | SRR330882(SRX091720) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR326282(GSM769512) Dicer mRNA was knocked down using siDicer, to. (cell line) | SRR363673(GSM830250) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | DRR000558(DRX000316) THP-1 whole cell RNA, after 3 day treatment w. (cell line) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577744(Rovira) total RNA. (breast) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR330914(SRX091752) tissue: normal skindisease state: normal. (skin) | SRR330858(SRX091696) tissue: skin psoriatic involveddisease state:. (skin) | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | SRR330859(SRX091697) tissue: skin psoriatic involveddisease state:. (skin) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR191547(GSM715657) 155genomic small RNA (size selected RNA from . (breast) | TAX577453(Rovira) total RNA. (breast) | SRR191488(GSM715598) 14genomic small RNA (size selected RNA from t. (breast) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR037939(GSM510477) 293cand5_rep1. (cell line) | DRR000555(DRX000313) THP-1 whole cell RNA, no treatment. (cell line) | SRR189785 | SRR191622(GSM715732) 175genomic small RNA (size selected RNA from . (breast) | SRR191613(GSM715723) 66genomic small RNA (size selected RNA from t. (breast) | GSM1105753INPUT(GSM1105753) small RNA sequencing data. (hela) | SRR040039(GSM532924) G531T. (cervix) | TAX577741(Rovira) total RNA. (breast) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR038856(GSM458539) D11. (cell line) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR060983(GSM569187) Human pre-germinal center B cell [09-001]. (cell line) | SRR038857(GSM458540) D20. (cell line) | SRR330880(SRX091718) tissue: skin psoriatic involveddisease state:. (skin) | SRR330903(SRX091741) tissue: skin psoriatic uninvolveddisease stat. (skin) | DRR000559(DRX000317) THP-1 whole cell RNA, no treatment. (cell line) | GSM450598(GSM450598) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330885(SRX091723) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR033711(GSM497056) GCB DLBCL (GCB110). (B cell) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR060984(GSM569188) Human plasma cell [09-001]. (cell line) | SRR330916(SRX091754) tissue: normal skindisease state: normal. (skin) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | SRR038861(GSM458544) MM466. (cell line) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR033717(GSM497062) Mentle Cell Lymphoma (MCL112). (B cell) | SRR191565(GSM715675) 92genomic small RNA (size selected RNA from t. (breast) | SRR189786 | SRR191434(GSM715544) 171genomic small RNA (size selected RNA from . (breast) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | DRR000557(DRX000315) THP-1 whole cell RNA, after 3 day treatment w. (cell line) | SRR444049(SRX128897) Sample 9cDNABarcode: AF-PP-334: ACG CTC TTC C. (skin) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR330899(SRX091737) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR189784 | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | DRR001488(DRX001042) Hela short nuclear cell fraction, LNA(+). (hela) | SRR095854(SRX039177) miRNA were isolated from FirstChoice Human Br. (brain) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................TCTGAGCCCTAGGATAAAA........................................... | 19 | 3.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................TCTCTGAGCCCTAGGATAA............................................. | 19 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................CCCAGCCCGCCGCCGGGGCGC | 21 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................CTCTGAGCCCTAGGACAA............................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................CGGCCGAGTACATCACCA.............................................................................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................GCCCGCCGCCGGGGCGC | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................................CAGCCCGCCGCCGGGGCCC | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................GGCCGAGTACATCAGTTAA............................................................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................GGCCGAGTACATCA................................................................................................................................. | 14 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................TCTCTGAGCCCTAGGAAAA............................................. | 19 | 0.25 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | |
| ..........................................................................................................CTCTGAGCCCTAGGAT............................................... | 16 | 8 | 0.25 | 0.25 | - | - | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.12 | - |
| .........................................................................................................TCTCTGAGCCCTAGGA................................................ | 16 | 4 | 0.25 | 0.25 | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................CTCTGAGCCCTAGGATCAA............................................ | 19 | 0.25 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.12 | - | - | |
| ............................CCGAGTACATCAG................................................................................................................................ | 13 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - | - | - | - | - |
| .....................................TCAGTGCTGAGAGGTG.................................................................................................................... | 16 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - | - | - |
| ..........................................................................................................CTCTGAGCCCTAGGATCAAA........................................... | 20 | 0.12 | 0.00 | - | - | - | - | - | - | - | - | - | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................CTCTGAGCCCTAGGATCACA........................................... | 20 | 0.12 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................GGGTGCCCGGGCCG......................................................................................... | 14 | 9 | 0.11 | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 |