| (2) AGO2.ip | (6) B-CELL | (8) BRAIN | (12) BREAST | (19) CELL-LINE | (2) CERVIX | (5) HEART | (3) HELA | (4) LIVER | (2) OTHER | (1) RRP40.ip | (22) SKIN | (1) TESTES | (3) UTERUS | (1) XRN.ip |
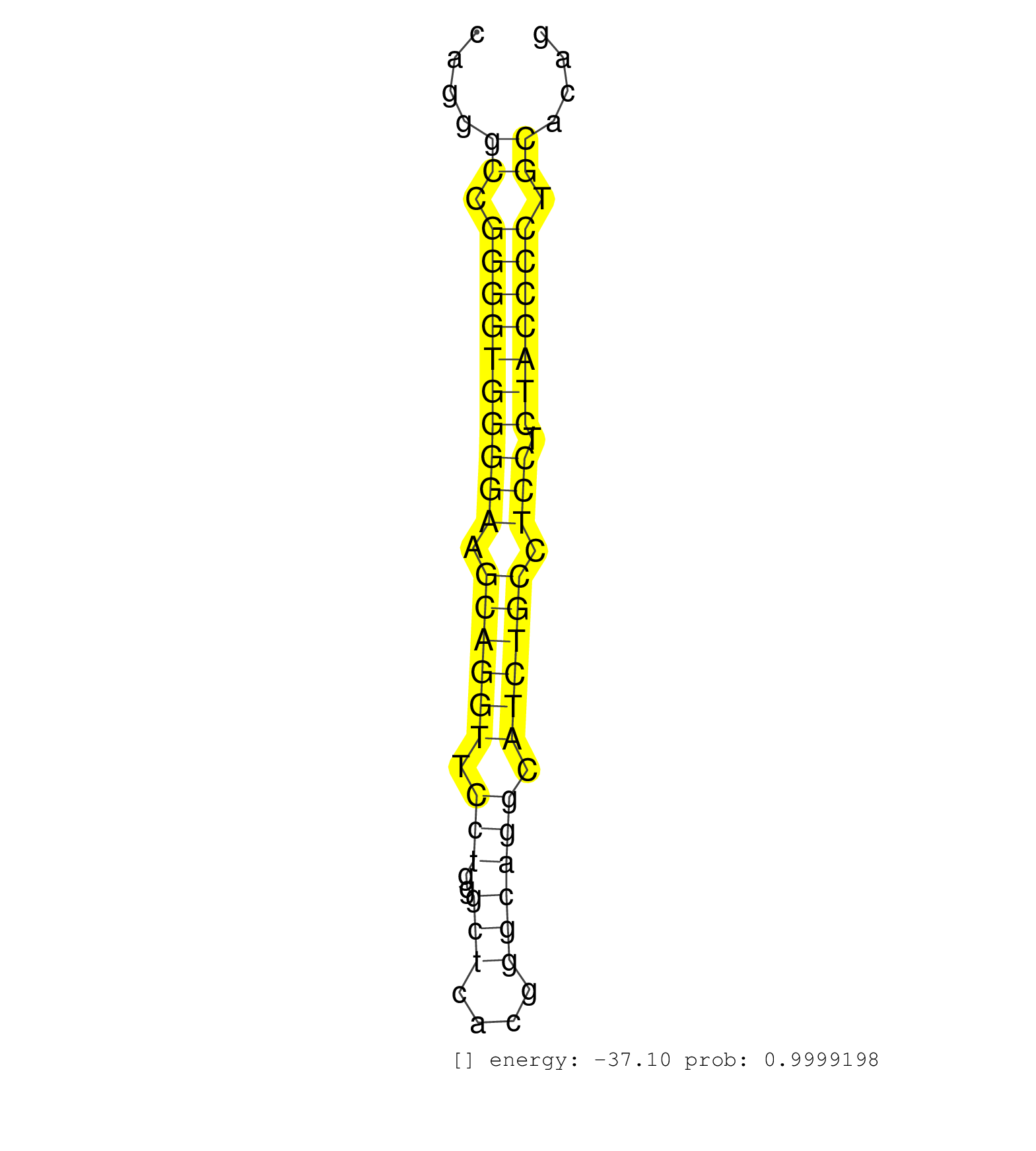
| CAGACATCCCAGCCTACTACATCAGTACTTTCAAGTTTGATCATGCACTTGTGAGTGTCCAGCGCCAGACCCCTCCAGGGCAGGGCCGGGGTGGGGAAGCAGGTTCCTGGGCTCACGGGCAGGCATCTGCCTCCTCTACCCCTGCACAGGTCCCCGAAGAGAACATCAATGGTGTCATCAGTGCCCTGAAGGTCAGCCA ....................................................................................((.((((((((((.((((((.(((..(((....)))))).)))))).))).))))))).))...................................................... ................................................................................81..................................................................149................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR314796(SRX084354) Total RNA, fractionated (15-30nt). (cell line) | SRR189782 | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR189787 | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR029131(GSM416760) MCF7. (cell line) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR189786 | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR330900(SRX091738) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR330920(SRX091758) tissue: normal skindisease state: normal. (skin) | SRR191606(GSM715716) 84genomic small RNA (size selected RNA from t. (breast) | SRR553576(SRX182782) source: Testis. (testes) | GSM450607(GSM450607) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | DRR001486(DRX001040) Hela long cytoplasmic cell fraction, LNA(+). (hela) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330903(SRX091741) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577580(Rovira) total RNA. (breast) | SRR330892(SRX091730) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | GSM1105753INPUT(GSM1105753) small RNA sequencing data. (hela) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | SRR037941(GSM510479) 293DroshaTN. (cell line) | SRR029132(GSM416761) MB-MDA231. (cell line) | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | TAX577453(Rovira) total RNA. (breast) | SRR040022(GSM532907) G575N. (cervix) | SRR037943(GSM510481) 293DcrTN. (cell line) | SRR343336 | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | SRR037935(GSM510473) 293cand3. (cell line) | SRR029126(GSM416755) 143B. (cell line) | SRR343333(GSM796036) KSHV (HHV8). (cell line) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330870(SRX091708) tissue: skin psoriatic involveddisease state:. (skin) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR444059(SRX128907) Sample 17cDNABarcode: AF-PP-339: ACG CTC TTC . (skin) | SRR033730(GSM497075) Lymphoblastic (ALL411). (B cell) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR191573(GSM715683) 68genomic small RNA (size selected RNA from t. (breast) | GSM450598(GSM450598) miRNA sequencing raw reads from post-mortem s. (brain) | SRR060169(GSM565979) 5-8F_cytoplasm. (cell line) | SRR330871(SRX091709) tissue: skin psoriatic involveddisease state:. (skin) | SRR191591(GSM715701) 53genomic small RNA (size selected RNA from t. (breast) | SRR330874(SRX091712) tissue: skin psoriatic involveddisease state:. (skin) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | SRR191590(GSM715700) 48genomic small RNA (size selected RNA from t. (breast) | SRR444049(SRX128897) Sample 9cDNABarcode: AF-PP-334: ACG CTC TTC C. (skin) | TAX577579(Rovira) total RNA. (breast) | SRR040016(GSM532901) G645N. (cervix) | SRR330876(SRX091714) tissue: skin psoriatic involveddisease state:. (skin) | SRR326281(GSM769511) Dicer mRNA was knocked down using siDicer, cy. (cell line) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | GSM450597(GSM450597) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | SRR039636(GSM518473) THP1_cyto_sRNAs. (cell line) | SRR191615(GSM715725) 94genomic small RNA (size selected RNA from t. (breast) | SRR191568(GSM715678) 51genomic small RNA (size selected RNA from t. (breast) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | TAX577588(Rovira) total RNA. (breast) | SRR191484(GSM715594) 12genomic small RNA (size selected RNA from t. (breast) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | DRR001488(DRX001042) Hela short nuclear cell fraction, LNA(+). (hela) | SRR444048(SRX128896) Sample 8cDNABarcode: AF-PP-333: ACG CTC TTC C. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................CATCTGCCTCCTCTACCCCTGC...................................................... | 22 | 2 | 13.50 | 13.50 | - | 2.00 | 1.50 | - | 0.50 | 0.50 | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | 0.50 | 0.50 | 0.50 | - | - | - | - | - | - | 0.50 | - | 0.50 | - | - | 0.50 | - | - | - | - | - |
| ...........................................................................................................................CATCTGCCTCCTCTACCCCTGCA..................................................... | 23 | 2 | 7.00 | 7.00 | 2.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | 0.50 | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | 0.50 | - | - | - | - | 0.50 | - | - | 0.50 | - | 0.50 | - | - | - | - |
| ...........................................................................................................................CATCTGCCTCCTCTACCCCTGCAC.................................................... | 24 | 2 | 6.00 | 6.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | 0.50 | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - |
| .....................................................................................CCGGGGTGGGGAAGCAGGTTC............................................................................................. | 21 | 2 | 5.50 | 5.50 | - | 2.00 | - | - | - | - | - | - | 2.50 | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................CATCTGCCTCCTCTACCCCTGCAT.................................................... | 24 | 2 | 5.00 | 7.00 | 3.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................CATCTGCCTCCTCTACCCCTGCATA................................................... | 25 | 2 | 3.00 | 7.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................CATCTGCCTCCTCTACCCCTG....................................................... | 21 | 2 | 3.00 | 3.00 | - | - | - | - | 1.00 | - | - | - | - | - | 1.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................CATCTGCCTCCTCTACCCCTGCAAT................................................... | 25 | 2 | 3.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................CATCTGCCTCCTCTACCCCTGCACTTT................................................. | 27 | 2 | 3.00 | 6.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................CATCTGCCTCCTCTACCCCTGCACT................................................... | 25 | 2 | 2.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................GCATCTGCCTCCTCTACCCCTG....................................................... | 22 | 2 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................CATCTGCCTCCTCTACCCCTGCACA................................................... | 25 | 2 | 2.00 | 2.00 | 1.00 | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................CCGGGGTGGGGAAGCAGGTTCT............................................................................................ | 22 | 2 | 2.00 | 5.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................CATCTGCCTCCTCTACCCCTGCAA.................................................... | 24 | 2 | 2.00 | 7.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................CGGGGTGGGGAAGCAGGT............................................................................................... | 18 | 2 | 1.50 | 1.50 | - | - | - | - | - | - | - | 1.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................GTGAGTGTCCAGCGCCAGACCCCTCCAGGGCAGGGCCGGGGTGGGGAAGCAGGTTCCTGGGCTCACGGGCAGGCATCTGCCTCCTCTACCCCTGCA..................................................... | 96 | 2 | 1.50 | 1.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................CATCTGCCTCCTCTACCCCTGCATT................................................... | 25 | 2 | 1.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................CCGGGGTGGGGAAGCAGGTTCC............................................................................................ | 22 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................TGCCTCCTCTACCCCTGCACA................................................... | 21 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................GCATCTGCCTCCTCTACCCCTGC...................................................... | 23 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................GCATCTGCCTCCTCTACCCCTGA...................................................... | 23 | 2 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................CGGGGTGGGGAAGCAGTTC.............................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................CATCTGCCTCCTCTACCCCTGCTT.................................................... | 24 | 2 | 1.00 | 13.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................CCGGGGTGGGGAAGCAGGTT.............................................................................................. | 20 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................CCGGGGTGGGGAAGCAGGGTCC............................................................................................ | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................CATCTGCCTCCTCTACCCCTGCAAA................................................... | 25 | 2 | 1.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...ACATCCCAGCCTACTACATCAGTACTTTCAAGT................................................................................................................................................................... | 33 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................TGCCTCCTCTACCCCTGCACAT.................................................. | 22 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................ATCTGCCTCCTCTACCCCTG....................................................... | 20 | 2 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................CATCTGCCTCCTCTACCCCTGCC..................................................... | 23 | 2 | 1.00 | 13.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................TCTGCCTCCTCTACCCCTGCAG.................................................... | 22 | 2 | 1.00 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................CGGGGTGGGGAAGCAGGTTC............................................................................................. | 20 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................CGGGGTGGGGAAGCAGGTTAT............................................................................................ | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................CATCTGCCTCCTCTACCCCTGA...................................................... | 22 | 2 | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........CAGCCTACTACATCAAG............................................................................................................................................................................. | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................CATCTGCCTCCTCTACCCCTGCATC................................................... | 25 | 2 | 1.00 | 7.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................CCGGGGTGGGGAAGCAGGTAA............................................................................................. | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................TGCCTCCTCTACCCCTGCACAGT................................................. | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................TCTGCCTCCTCTACCCCTGC...................................................... | 20 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................TCTGCCTCCTCTACCCCTGCACA................................................... | 23 | 2 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................CCGGGGTGGGGAAGCAGGTTAA............................................................................................ | 22 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................CGAAGAGAACATCAATG............................ | 17 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................GCATCTGCCTCCTCTACCC.......................................................... | 19 | 2 | 0.50 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................GCCGGGGTGGGGAAGCAGGTTCCT........................................................................................... | 24 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................TCCCCGAAGAGAACATCAATGGTGTCATC.................... | 29 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................TGGTGTCATCAGTGCCCTGAAGGTCAGC.. | 28 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................CGGGGTGGGGAAGCAGGTTCC............................................................................................ | 21 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................TCTGCCTCCTCTACCCCTGCA..................................................... | 21 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................ATCAGTACTTTCAAGTTTG................................................................................................................................................................ | 19 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................TCTGCCTCCTCTACCCCT........................................................ | 18 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - |
| ........................................................................................................................................................................ATGGTGTCATCAGTGCCC............. | 18 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................CGGGGTGGGGAAGCAGGATC............................................................................................. | 20 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - |
| ......................................................................................CGGGGTGGGGAAGCAGG................................................................................................ | 17 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................CTGCCTCCTCTACCCCT........................................................ | 17 | 9 | 0.11 | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 | - |
| CAGACATCCCAGCCTACTACATCAGTACTTTCAAGTTTGATCATGCACTTGTGAGTGTCCAGCGCCAGACCCCTCCAGGGCAGGGCCGGGGTGGGGAAGCAGGTTCCTGGGCTCACGGGCAGGCATCTGCCTCCTCTACCCCTGCACAGGTCCCCGAAGAGAACATCAATGGTGTCATCAGTGCCCTGAAGGTCAGCCA ....................................................................................((.((((((((((.((((((.(((..(((....)))))).)))))).))).))))))).))...................................................... ................................................................................81..................................................................149................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR314796(SRX084354) Total RNA, fractionated (15-30nt). (cell line) | SRR189782 | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR189787 | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR029131(GSM416760) MCF7. (cell line) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR189786 | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR330900(SRX091738) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR330920(SRX091758) tissue: normal skindisease state: normal. (skin) | SRR191606(GSM715716) 84genomic small RNA (size selected RNA from t. (breast) | SRR553576(SRX182782) source: Testis. (testes) | GSM450607(GSM450607) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | DRR001486(DRX001040) Hela long cytoplasmic cell fraction, LNA(+). (hela) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330903(SRX091741) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577580(Rovira) total RNA. (breast) | SRR330892(SRX091730) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | GSM1105753INPUT(GSM1105753) small RNA sequencing data. (hela) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | SRR037941(GSM510479) 293DroshaTN. (cell line) | SRR029132(GSM416761) MB-MDA231. (cell line) | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | TAX577453(Rovira) total RNA. (breast) | SRR040022(GSM532907) G575N. (cervix) | SRR037943(GSM510481) 293DcrTN. (cell line) | SRR343336 | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | SRR037935(GSM510473) 293cand3. (cell line) | SRR029126(GSM416755) 143B. (cell line) | SRR343333(GSM796036) KSHV (HHV8). (cell line) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330870(SRX091708) tissue: skin psoriatic involveddisease state:. (skin) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR444059(SRX128907) Sample 17cDNABarcode: AF-PP-339: ACG CTC TTC . (skin) | SRR033730(GSM497075) Lymphoblastic (ALL411). (B cell) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR191573(GSM715683) 68genomic small RNA (size selected RNA from t. (breast) | GSM450598(GSM450598) miRNA sequencing raw reads from post-mortem s. (brain) | SRR060169(GSM565979) 5-8F_cytoplasm. (cell line) | SRR330871(SRX091709) tissue: skin psoriatic involveddisease state:. (skin) | SRR191591(GSM715701) 53genomic small RNA (size selected RNA from t. (breast) | SRR330874(SRX091712) tissue: skin psoriatic involveddisease state:. (skin) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | SRR191590(GSM715700) 48genomic small RNA (size selected RNA from t. (breast) | SRR444049(SRX128897) Sample 9cDNABarcode: AF-PP-334: ACG CTC TTC C. (skin) | TAX577579(Rovira) total RNA. (breast) | SRR040016(GSM532901) G645N. (cervix) | SRR330876(SRX091714) tissue: skin psoriatic involveddisease state:. (skin) | SRR326281(GSM769511) Dicer mRNA was knocked down using siDicer, cy. (cell line) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | GSM450597(GSM450597) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | SRR039636(GSM518473) THP1_cyto_sRNAs. (cell line) | SRR191615(GSM715725) 94genomic small RNA (size selected RNA from t. (breast) | SRR191568(GSM715678) 51genomic small RNA (size selected RNA from t. (breast) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | TAX577588(Rovira) total RNA. (breast) | SRR191484(GSM715594) 12genomic small RNA (size selected RNA from t. (breast) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | DRR001488(DRX001042) Hela short nuclear cell fraction, LNA(+). (hela) | SRR444048(SRX128896) Sample 8cDNABarcode: AF-PP-333: ACG CTC TTC C. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............CCTACTACATCAGTAATC......................................................................................................................................................................... | 18 | 5.00 | 0.00 | - | - | - | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........CAGCCTACTACATCATGGT........................................................................................................................................................................... | 19 | 4.00 | 0.00 | - | - | 3.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........CAGCCTACTACATCATGTG........................................................................................................................................................................... | 19 | 4.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........CAGCCTACTACATCATAT............................................................................................................................................................................ | 18 | 3.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........CAGCCTACTACATCATGG............................................................................................................................................................................ | 18 | 2.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........CAGCCTACTACATCATGTT........................................................................................................................................................................... | 19 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........CAGCCTACTACATCACATG........................................................................................................................................................................... | 19 | 2.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........CAGCCTACTACATCACG............................................................................................................................................................................. | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........CAGCCTACTACATCATATG........................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........CAGCCTACTACATCATATA........................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........CCAGCCTACTACATCATTT............................................................................................................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........CAGCCTACTACATCATAAC........................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........CCAGCCTACTACATCATT............................................................................................................................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........CAGCCTACTACATCATGC............................................................................................................................................................................ | 18 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......CCCAGCCTACTACATCATTGT........................................................................................................................................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........CAGCCTACTACATCATAA............................................................................................................................................................................ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........CAGCCTACTACATCAGTGTG.......................................................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................AGTTTGATCATGCACTTATAT................................................................................................................................................. | 21 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........CAGCCTACTACATCACTTG........................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........CAGCCTACTACATCATTC............................................................................................................................................................................ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......TCCCAGCCTACTACATCAAGGT........................................................................................................................................................................... | 22 | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........CCAGCCTACTACATCAATAG........................................................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................TCCTCTACCCCTGCACAC.................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......CCCAGCCTACTACATCATATG........................................................................................................................................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......TCCCAGCCTACTACATCAAGTT........................................................................................................................................................................... | 22 | 1.00 | 0.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......CCCAGCCTACTACATCATCGT........................................................................................................................................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........CCAGCCTACTACATCA............................................................................................................................................................................... | 16 | 2 | 0.50 | 0.50 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........GCCTACTACATCAGTACTTTCAAGTTTGA............................................................................................................................................................... | 29 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................CTCCTCTACCCCTGCACA................................................... | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................TACATCAGTACTTTCAAGTT.................................................................................................................................................................. | 20 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................GCCTCCTCTACCCCTG....................................................... | 16 | 9 | 0.11 | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................TCCAGCGCCAGACC................................................................................................................................ | 14 | 10 | 0.10 | 0.10 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.10 |