| (1) AGO1.ip | (1) AGO2.ip | (1) AGO3.ip | (1) BRAIN | (1) BREAST | (3) CELL-LINE | (3) HEART | (3) HELA | (12) LIVER | (2) OTHER | (9) SKIN |
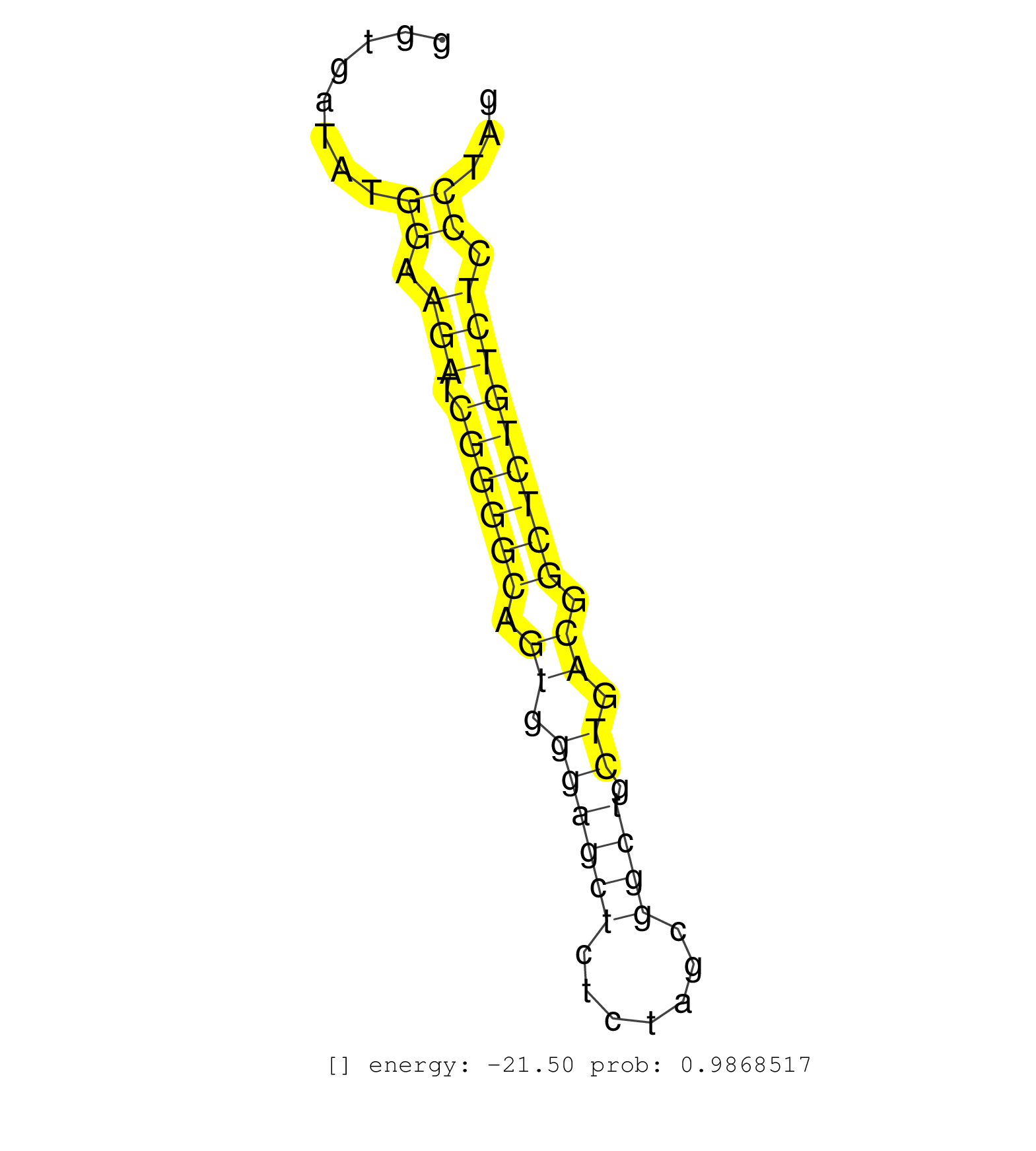
| TCTCCTGGGGGAAGTTCTTAGAGGCCAATGGAGTGATCTTGGCCTTTCAAGCAGCAAGCTAGGTCCACTAGGGAAAGGTGACAGAGGTGTGAACAGACAGACCCTCTGTCCTGCTGACCACTTGACCCTGGAGAGGGGTGATATGGAAGATCGGGGCAGTGGGAGCTCTCTAGCGGCTGCTGACGGCTCTGTCTCCCTAGGCGGAAGGCAGGTGGCCGCCGCCGGAGCAATGGTGCAAACAGCTCTTCTC ................................................................................................................................................((.(((.((((((.((.((((((.......)))).)).)).))))))))).))..................................................... ........................................................................................................................................137............................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | GSM1105748AGO1(GSM1105748) small RNA sequencing data. (ago1 hela) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR330923(SRX091761) tissue: normal skindisease state: normal. (skin) | SRR039612(GSM531975) Human Normal Liver Tissue Sample 2. (liver) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR330913(SRX091751) tissue: normal skindisease state: normal. (skin) | SRR444061(SRX128909) Sample 19cDNABarcode: AF-PP-341: ACG CTC TTC . (skin) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | GSM1105749AGO2(GSM1105749) small RNA sequencing data. (ago2 hela) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | SRR189786 | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | TAX577739(Rovira) total RNA. (breast) | SRR037937(GSM510475) 293cand2. (cell line) | SRR330881(SRX091719) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM1105750AGO3(GSM1105750) small RNA sequencing data. (ago3 hela) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR039611(GSM531974) Human Normal Liver Tissue Sample 1. (liver) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM956925Paz8D5(GSM956925) cell line: HEK293 cell linepassages: 15-20ip . (cell line) | GSM450599(GSM450599) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330895(SRX091733) tissue: skin psoriatic uninvolveddisease stat. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................TATGGAAGATCGGGGCAG........................................................................................... | 18 | 1 | 10.00 | 10.00 | 1.00 | - | 1.00 | - | 2.00 | - | 2.00 | - | - | 2.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TATGGAAGATCGGGGCAGT.......................................................................................... | 19 | 1 | 7.00 | 7.00 | 2.00 | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .............................................................................................................................................TATGGAAGATCGGGGCAGTGGGA...................................................................................... | 23 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TATGGAAGATCGGGGCAGTGG........................................................................................ | 21 | 1 | 4.00 | 4.00 | - | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .............................................................................................................................................TATGGAAGATCGGGGCAGTAA........................................................................................ | 21 | 1 | 4.00 | 7.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TATGGAAGATCGGGGCAGTGGG....................................................................................... | 22 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................CTGACGGCTCTGTCTCCCTA................................................... | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................ATATGGAAGATCGGGGCAG........................................................................................... | 19 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TATGGAAGATCGGGGCATTAT........................................................................................ | 21 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................TATGGAAGATCGGGGCAGTGGGATT.................................................................................... | 25 | 1 | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TGACGGCTCTGTCTCCCTAGTA................................................ | 22 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................................CGGGGCAGTGGGAGCTCTCTAGCGGC......................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TATGGAAGATCGGGGCAGTGGA....................................................................................... | 22 | 1 | 1.00 | 4.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TATGGAAGATCGGGGCAGTGGGAA..................................................................................... | 24 | 1 | 1.00 | 4.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TATGGAAGATCGGGGCAGTAAT....................................................................................... | 22 | 1 | 1.00 | 7.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TATGGAAGATCGGGGCAGTGGGAT..................................................................................... | 24 | 1 | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................CCTAGGCGGAAGGCAGGTGGC.................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TGACGGCTCTGTCTCCCTAGT................................................. | 21 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................TGACGGCTCTGTCTCCCTAGA................................................. | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | |
| .............................................................................................................................................TATGGAAGATCGGGGCAGTAAGA...................................................................................... | 23 | 1 | 1.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................ATGGAAGATCGGGGCAGTGGGT...................................................................................... | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | |
| .............................................................................................................................................TATGGAAGATCGGGGCAGG.......................................................................................... | 19 | 1 | 1.00 | 10.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................CTGACGGCTCTGTCTCCCTAGA................................................. | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................AGAGGTGTGAACAGACAGACCCTC................................................................................................................................................ | 24 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TGACGGCTCTGTCTCCATA................................................... | 19 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................TATGGAAGATCGGGGCAGTGAAA...................................................................................... | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................GGGAAAGGTGACAGAGGTGTG............................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................GAAGATCGGGGCAGTGG........................................................................................ | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TGCTGACGGCTCTGTCCTTC..................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................AGAGGTGTGAACAGACAGACCTTCT............................................................................................................................................... | 25 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................TATGGAAGATCGGGGCAAAAA........................................................................................ | 21 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................TATGGAAGATCGGGGCAGTGGGAAA.................................................................................... | 25 | 1 | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................GCAGGTGGCCGCCGCCGGAGCA..................... | 22 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TATGGAAGATCGGGGCAGTGA........................................................................................ | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................AGGGAAAGGTGACAGAGGTGTGAAC............................................................................................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TATGGAAGATCGGGGCAGTGGAAAT.................................................................................... | 25 | 1 | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TATGGAAGATCGGGGCAGTGGAAA..................................................................................... | 24 | 1 | 1.00 | 4.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................AGGGAAAGGTGACAGAGGTGT................................................................................................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................CGGGGCAGTGGGAGCT................................................................................... | 16 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 |
| TCTCCTGGGGGAAGTTCTTAGAGGCCAATGGAGTGATCTTGGCCTTTCAAGCAGCAAGCTAGGTCCACTAGGGAAAGGTGACAGAGGTGTGAACAGACAGACCCTCTGTCCTGCTGACCACTTGACCCTGGAGAGGGGTGATATGGAAGATCGGGGCAGTGGGAGCTCTCTAGCGGCTGCTGACGGCTCTGTCTCCCTAGGCGGAAGGCAGGTGGCCGCCGCCGGAGCAATGGTGCAAACAGCTCTTCTC ................................................................................................................................................((.(((.((((((.((.((((((.......)))).)).)).))))))))).))..................................................... ........................................................................................................................................137............................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | GSM1105748AGO1(GSM1105748) small RNA sequencing data. (ago1 hela) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR330923(SRX091761) tissue: normal skindisease state: normal. (skin) | SRR039612(GSM531975) Human Normal Liver Tissue Sample 2. (liver) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR330913(SRX091751) tissue: normal skindisease state: normal. (skin) | SRR444061(SRX128909) Sample 19cDNABarcode: AF-PP-341: ACG CTC TTC . (skin) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | GSM1105749AGO2(GSM1105749) small RNA sequencing data. (ago2 hela) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | SRR189786 | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | TAX577739(Rovira) total RNA. (breast) | SRR037937(GSM510475) 293cand2. (cell line) | SRR330881(SRX091719) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM1105750AGO3(GSM1105750) small RNA sequencing data. (ago3 hela) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR039611(GSM531974) Human Normal Liver Tissue Sample 1. (liver) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM956925Paz8D5(GSM956925) cell line: HEK293 cell linepassages: 15-20ip . (cell line) | GSM450599(GSM450599) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330895(SRX091733) tissue: skin psoriatic uninvolveddisease stat. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................................................................................CAGGTGGCCGCCGCCGGGG....................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............AGTTCTTAGAGGCCAACTG........................................................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | |
| ..................................................................................................................................................................................................................GGTGGCCGCCGCCGGAGCC..................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | |
| .............................................................................................CAGACAGACCCTCTGTAGCG......................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................................GGGGCAGTGGGAGCTCTACC.............................................................................. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................CTCTGTCCTGCTGACCAG................................................................................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |