| (5) BREAST | (6) CELL-LINE | (8) CERVIX | (2) FIBROBLAST | (9) HEART | (1) HELA | (1) LIVER | (2) OTHER | (30) SKIN | (1) TESTES | (2) UTERUS |
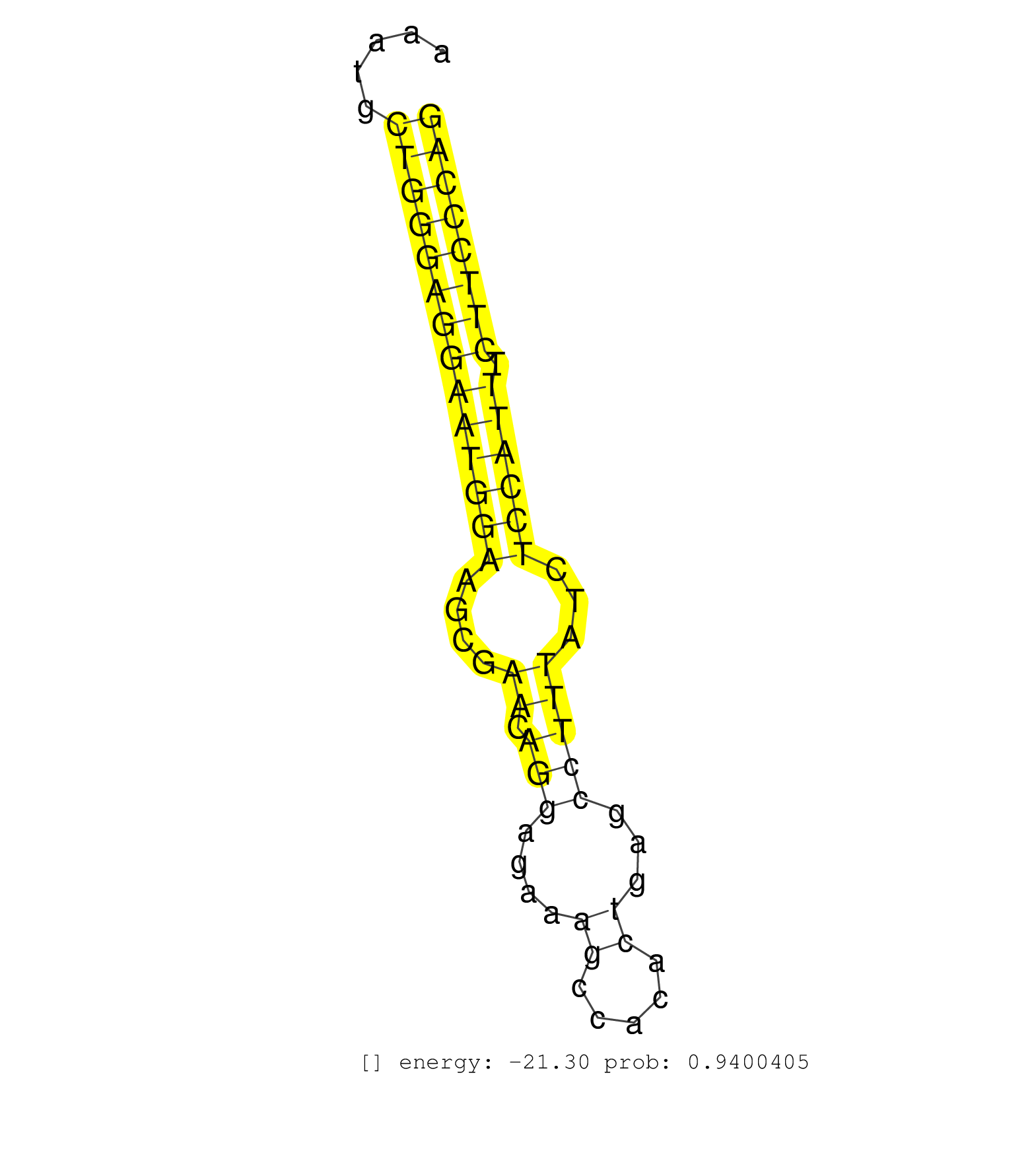
| ATCCAGGGACTCCCAAACTCTAAAAAGTAGCATTTTTTACTTCAAATCTAAAAGGTGGAGCTAATTTTGTGAACAATAGCTTCAATCTCCATTGCTAAAGAAGGCATGGCCTGAGAATTTTGCAACTTTGCAAAATGCTGGGAGGAATGGAAGCGAACAGGAGAAAGCCACACTGAGCCTTTATCTCCATTTCTTCCCAGTACTGAGCCAGGCTGAGAGCTGGCAGCGGCTGCACAATGAGACCCGCACT .........................................................................................................................................((((((((((((((....((.(((....((.....))...)))))...)))))).)))))))).................................................. ....................................................................................................................................133................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR189782 | SRR343335 | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | SRR330900(SRX091738) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR040031(GSM532916) G013T. (cervix) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330870(SRX091708) tissue: skin psoriatic involveddisease state:. (skin) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR040018(GSM532903) G701N. (cervix) | TAX577739(Rovira) total RNA. (breast) | SRR330881(SRX091719) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330920(SRX091758) tissue: normal skindisease state: normal. (skin) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR330913(SRX091751) tissue: normal skindisease state: normal. (skin) | SRR189784 | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR040016(GSM532901) G645N. (cervix) | GSM532876(GSM532876) G547T. (cervix) | SRR330876(SRX091714) tissue: skin psoriatic involveddisease state:. (skin) | SRR330872(SRX091710) tissue: skin psoriatic involveddisease state:. (skin) | SRR330904(SRX091742) tissue: normal skindisease state: normal. (skin) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR444049(SRX128897) Sample 9cDNABarcode: AF-PP-334: ACG CTC TTC C. (skin) | TAX577590(Rovira) total RNA. (breast) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR330865(SRX091703) tissue: skin psoriatic involveddisease state:. (skin) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR553576(SRX182782) source: Testis. (testes) | SRR330912(SRX091750) tissue: normal skindisease state: normal. (skin) | SRR343334 | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330916(SRX091754) tissue: normal skindisease state: normal. (skin) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR038862(GSM458545) MM472. (cell line) | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | SRR330923(SRX091761) tissue: normal skindisease state: normal. (skin) | SRR189783 | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | TAX577744(Rovira) total RNA. (breast) | SRR363674(GSM830251) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR444052(SRX128900) Sample 12cDNABarcode: AF-PP-340: ACG CTC TTC . (skin) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR191417(GSM715527) 39genomic small RNA (size selected RNA from t. (breast) | SRR191603(GSM715713) 71genomic small RNA (size selected RNA from t. (breast) | SRR363675(GSM830252) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR040010(GSM532895) G529N. (cervix) | SRR553574(SRX182780) source: Heart. (Heart) | SRR040012(GSM532897) G648N. (cervix) | SRR029124(GSM416753) HeLa. (hela) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR040022(GSM532907) G575N. (cervix) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR040008(GSM532893) G727N. (cervix) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................................................TTTATCTCCATTTCTTCCCAGT................................................. | 22 | 1 | 14.00 | 14.00 | 1.00 | - | - | 2.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | 1.00 | 1.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - |
| ...................................................................................................................................................................................TTTATCTCCATTTCTTCCCATT................................................. | 22 | 9.00 | 0.00 | - | 1.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | |
| .........................................................................................................................................CTGGGAGGAATGGAAGCGAACAG.......................................................................................... | 23 | 1 | 7.00 | 7.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TTATCTCCATTTCTTCCCATTT................................................ | 22 | 5.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................TTTATCTCCATTTCTTCCCAAT................................................. | 22 | 4.00 | 0.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | |
| .........................................................................................................................................CTGGGAGGAATGGAAGCGAACAGG......................................................................................... | 24 | 1 | 4.00 | 4.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TTATCTCCATTTCTTCCCAGT................................................. | 21 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................CTGGGAGGAATGGAAGCGAACAGA......................................................................................... | 24 | 1 | 3.00 | 7.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TTTATCTCCATTTCTTCCCAGTT................................................ | 23 | 1 | 2.00 | 14.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................CTGGGAGGAATGGAAGCGA.............................................................................................. | 19 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TTTATCTCCATTTCTTCCCAGA................................................. | 22 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................................CTGGGAGGAATGGAAGCGAAC............................................................................................ | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TTATCTCCATTTCTTCCCAGTT................................................ | 22 | 1 | 2.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................CTGGGAGGAATGGAAGCGAACAGGT........................................................................................ | 25 | 1 | 2.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TTTATCTCCATTTCTTCCCATTT................................................ | 23 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................TTTATCTCCATTTCTTCCCAAA................................................. | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................TTTATCTCCATTTCTTCCCATAT................................................ | 23 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................................CTGGGAGGAATGGAAGCGAGAAA.......................................................................................... | 23 | 1 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TTTATCTCCATTTCTTCCC.................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .................................................................................................................................................................................................................AGGCTGAGAGCTGGCAGC....................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................CTGGGAGGAATGGAAGCGAACAGTT........................................................................................ | 25 | 1 | 1.00 | 7.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................TAGCATTTTTTACTTCACTTG.......................................................................................................................................................................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................TTATCTCCATTTCTTCCCAGACT............................................... | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................TTATCTCCATTTCTTCCCAGTTT............................................... | 23 | 1 | 1.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................CTGGGAGGAATGGAAGCGAACAGGA........................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................CTGGGAGGAATGGAAGCGAACAGGTTT...................................................................................... | 27 | 1 | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................CCAGGCTGAGAGCTGGCAGCGGC.................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................GCTGGGAGGAATGGAAGCGAACAGG......................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TTATCTCCATTTCTTCCCAAAT................................................ | 22 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................................................................................................ACTGAGCCAGGCTGAGAGCT............................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................CTGGGAGGAATGGAAGCGAAA............................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................CAGGCTGAGAGCTGGCAGC....................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................CTGGGAGGAATGGAAGCGAAGAA.......................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................CTGGGAGGAATGGAAGCGAA............................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TTATCTCCATTTCTTCCCAGAA................................................ | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................CTTTATCTCCATTTCTTCCCATTT................................................ | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................................CTGGGAGGAATGGAAGCGAACA........................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...................................................................................................................................................................................TTTATCTCCATTTCTTCCCATA................................................. | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................................CTGGGAGGAATGGAAGCGAACAGT......................................................................................... | 24 | 1 | 1.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................AGGCTGAGAGCTGGCAGCGGCTGC................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................TACTGAGCCAGGCTGAGAGCTGGCAGC....................... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................GCTGGGAGGAATGGAAGCGAACAG.......................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................CAGGCTGAGAGCTGGCAGCGGCT................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TTTATCTCCATTTCTTCCCAGTTT............................................... | 24 | 1 | 1.00 | 14.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................CAAAATGCTGGGAGGAATGG.................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................GCTGGGAGGAATGGAAGCGAAC............................................................................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TTATCTCCATTTCTTCCCAGTCC............................................... | 23 | 1 | 1.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................CAGGCTGAGAGCTGGCAGCGGCTGCA................ | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................................................................TACTGAGCCAGGCTGAGAGCT............................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................AGGCTGAGAGCTGGCAG........................ | 17 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - |
| .........................................................................................................................................CTGGGAGGAATGGAAGCG............................................................................................... | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 |
| ATCCAGGGACTCCCAAACTCTAAAAAGTAGCATTTTTTACTTCAAATCTAAAAGGTGGAGCTAATTTTGTGAACAATAGCTTCAATCTCCATTGCTAAAGAAGGCATGGCCTGAGAATTTTGCAACTTTGCAAAATGCTGGGAGGAATGGAAGCGAACAGGAGAAAGCCACACTGAGCCTTTATCTCCATTTCTTCCCAGTACTGAGCCAGGCTGAGAGCTGGCAGCGGCTGCACAATGAGACCCGCACT .........................................................................................................................................((((((((((((((....((.(((....((.....))...)))))...)))))).)))))))).................................................. ....................................................................................................................................133................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR189782 | SRR343335 | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | SRR330900(SRX091738) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR040031(GSM532916) G013T. (cervix) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330870(SRX091708) tissue: skin psoriatic involveddisease state:. (skin) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR040018(GSM532903) G701N. (cervix) | TAX577739(Rovira) total RNA. (breast) | SRR330881(SRX091719) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330920(SRX091758) tissue: normal skindisease state: normal. (skin) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR330913(SRX091751) tissue: normal skindisease state: normal. (skin) | SRR189784 | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR040016(GSM532901) G645N. (cervix) | GSM532876(GSM532876) G547T. (cervix) | SRR330876(SRX091714) tissue: skin psoriatic involveddisease state:. (skin) | SRR330872(SRX091710) tissue: skin psoriatic involveddisease state:. (skin) | SRR330904(SRX091742) tissue: normal skindisease state: normal. (skin) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR444049(SRX128897) Sample 9cDNABarcode: AF-PP-334: ACG CTC TTC C. (skin) | TAX577590(Rovira) total RNA. (breast) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR330865(SRX091703) tissue: skin psoriatic involveddisease state:. (skin) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR553576(SRX182782) source: Testis. (testes) | SRR330912(SRX091750) tissue: normal skindisease state: normal. (skin) | SRR343334 | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330916(SRX091754) tissue: normal skindisease state: normal. (skin) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR038862(GSM458545) MM472. (cell line) | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | SRR330923(SRX091761) tissue: normal skindisease state: normal. (skin) | SRR189783 | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | TAX577744(Rovira) total RNA. (breast) | SRR363674(GSM830251) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR444052(SRX128900) Sample 12cDNABarcode: AF-PP-340: ACG CTC TTC . (skin) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR191417(GSM715527) 39genomic small RNA (size selected RNA from t. (breast) | SRR191603(GSM715713) 71genomic small RNA (size selected RNA from t. (breast) | SRR363675(GSM830252) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR040010(GSM532895) G529N. (cervix) | SRR553574(SRX182780) source: Heart. (Heart) | SRR040012(GSM532897) G648N. (cervix) | SRR029124(GSM416753) HeLa. (hela) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR040022(GSM532907) G575N. (cervix) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR040008(GSM532893) G727N. (cervix) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................AAAATGCTGGGAGGAAATG.................................................................................................... | 19 | 5.00 | 0.00 | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................................................................................AGGCTGAGAGCTGGCACTC...................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................................TTCTTCCCAGTACTGGAT.......................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................GGGAGGAATGGAAGCCCCT............................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................................................................................................TGAGAGCTGGCAGCGG..................... | 16 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................AAAAGTAGCATTTTTTAG.................................................................................................................................................................................................................. | 18 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................GTAGCATTTTTTACTTCAGATT.......................................................................................................................................................................................................... | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................AAGCGAACAGGAGAATTT.................................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................ACAATAGCTTCAATCTCATA.............................................................................................................................................................. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................GAGCCTTTATCTCCACTGA......................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |