| (2) AGO1.ip | (1) AGO1.ip OTHER.mut | (4) AGO2.ip | (1) AGO3.ip | (14) B-CELL | (1) BRAIN | (4) BREAST | (36) CELL-LINE | (2) CERVIX | (2) HELA | (1) LIVER | (1) OTHER | (1) RRP40.ip | (3) SKIN | (1) UTERUS | (1) XRN.ip |
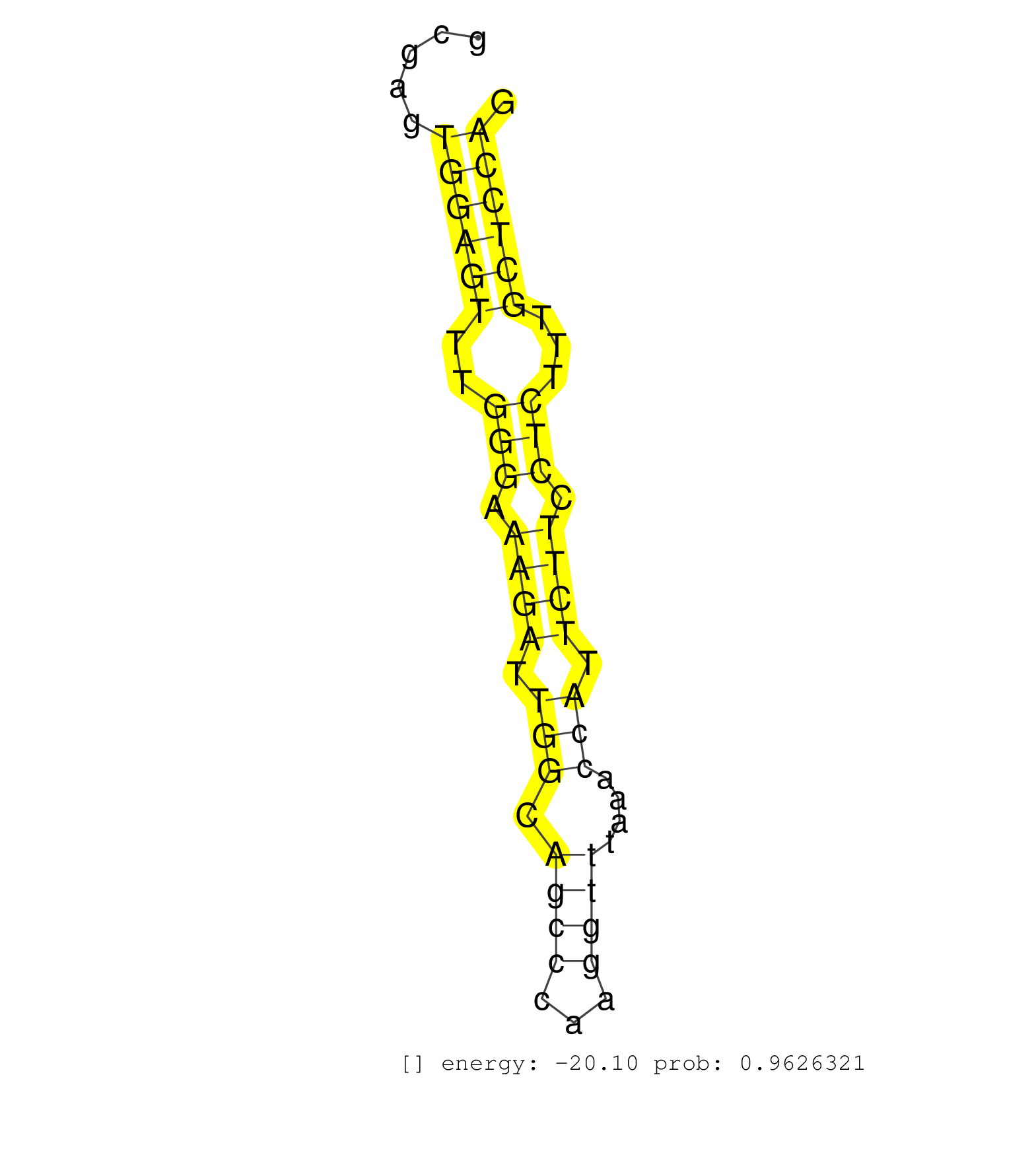
| TGGCTGCAGTGTCTGGCAGATTGGAAAAGGGATGATATCAGAAGGAGACAGACGGGTCAGAACATTCAGGAAAACTTCAAGTGAGACATAGAGCTTTGCCAGTAAAATCAGAAAGTGAAAAAACAGTCTTGAAAGGGGCGAGTGGAGTTTGGGAAAGATTGGCAGCCCAAGGTTTAAACCATTCTTCCTCTTTGCTCCAGGTGACCTCCTTGTTGCAGTTGGTTCATTCCTGCAGTGAGCAGTCTCCTCA ..............................................................................................................................................((((((..(((.((((.(((.((((...))))....))).)))).)))...))))))................................................... .........................................................................................................................................138...........................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR037938(GSM510476) 293Red. (cell line) | SRR207114(GSM721076) IP against AGO 1 & 2, RRP40 knockdown. (ago1/2 RRP40 cell line) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR038861(GSM458544) MM466. (cell line) | SRR033730(GSM497075) Lymphoblastic (ALL411). (B cell) | SRR033729(GSM497074) splenic MZL (Splenic414). (B cell) | SRR207116(GSM721078) Nuclear RNA. (cell line) | GSM1105748AGO1(GSM1105748) small RNA sequencing data. (ago1 hela) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | TAX577739(Rovira) total RNA. (breast) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | TAX577743(Rovira) total RNA. (breast) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR060982(GSM569186) Human centrocyte [09-001]. (cell line) | SRR038859(GSM458542) MM386. (cell line) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR326281(GSM769511) Dicer mRNA was knocked down using siDicer, cy. (cell line) | DRR000555(DRX000313) THP-1 whole cell RNA, no treatment. (cell line) | SRR038856(GSM458539) D11. (cell line) | SRR060983(GSM569187) Human pre-germinal center B cell [09-001]. (cell line) | SRR038858(GSM458541) MEL202. (cell line) | SRR040028(GSM532913) G026N. (cervix) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR033719(GSM497064) 6hr Activated B cell line (ABC158). (B cell) | SRR033723(GSM497068) L1236 cell line (L1236). (B cell) | SRR033717(GSM497062) Mentle Cell Lymphoma (MCL112). (B cell) | SRR189786 | SRR033716(GSM497061) Mentle Cell Lymphoma (MCL114). (B cell) | SRR330894(SRX091732) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033711(GSM497056) GCB DLBCL (GCB110). (B cell) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | DRR000562(DRX000320) Isolation of RNA following immunoprecipitatio. (ago3 cell line) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189787 | SRR033720(GSM497065) EBV activated B cell line (EBV159). (B cell) | SRR039193(GSM494812) HL60 cell line is derived from acute promyelo. (cell line) | DRR000558(DRX000316) THP-1 whole cell RNA, after 3 day treatment w. (cell line) | GSM1105749AGO2(GSM1105749) small RNA sequencing data. (ago2 hela) | SRR033732(GSM497077) bjab cell line (bjab103). (B cell) | SRR189782 | SRR060985(GSM569189) Human naive B cell [09-001]. (cell line) | TAX577453(Rovira) total RNA. (breast) | DRR000557(DRX000315) THP-1 whole cell RNA, after 3 day treatment w. (cell line) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR039637(GSM518474) THP1_total_sRNAs. (cell line) | SRR037933(GSM510471) 293cand4_rep2. (cell line) | GSM416733(GSM416733) HEK293. (cell line) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR033715(GSM497060) Mantle Cell Lymphoma (Mino122). (B cell) | SRR037931(GSM510469) 293GFP. (cell line) | SRR029126(GSM416755) 143B. (cell line) | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | DRR000561(DRX000319) Isolation of RNA following immunoprecipitatio. (ago2 cell line) | SRR037936(GSM510474) 293cand1. (cell line) | GSM450603(GSM450603) miRNA sequencing raw reads from post-mortem s. (brain) | SRR037934(GSM510472) 293cand4_rep3. (cell line) | GSM532883(GSM532883) G871N. (cervix) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................TGGAGTTTGGGAAAGATTGGCA...................................................................................... | 22 | 1 | 48.00 | 48.00 | 13.00 | - | 8.00 | 2.00 | 3.00 | 1.00 | - | 3.00 | - | - | - | 1.00 | - | 2.00 | 1.00 | 1.00 | - | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | 1.00 | - | 1.00 | 1.00 | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................TGGAGTTTGGGAAAGATTGGC....................................................................................... | 21 | 2 | 10.50 | 10.50 | 3.50 | - | - | - | 0.50 | 0.50 | - | - | - | 0.50 | - | 1.00 | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | 0.50 | - | - | - | - | - | - | - | 0.50 | 0.50 | 0.50 | 0.50 | 0.50 | - | - | 0.50 | - | - |
| ....................................................................................................................................................................................ATTCTTCCTCTTTGCTCCAGA................................................. | 21 | 10.00 | 0.00 | 10.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................ATTCTTCCTCTTTGCTCCAGAAT............................................... | 23 | 10.00 | 0.00 | - | 10.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................TGGAGTTTGGGAAAGATTGGCAA..................................................................................... | 23 | 1 | 8.00 | 48.00 | 2.00 | - | - | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................GTGGAGTTTGGGAAAGATTGG........................................................................................ | 21 | 2 | 8.00 | 8.00 | 4.00 | - | - | - | 0.50 | - | 1.50 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................TGGAGTTTGGGAAAGATTGGCAT..................................................................................... | 23 | 1 | 5.00 | 48.00 | 2.00 | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................AGTGGAGTTTGGGAAAGATTGG........................................................................................ | 22 | 2 | 5.00 | 5.00 | - | - | - | - | - | - | 1.00 | - | 1.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | 0.50 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................TGGAGTTTGGGAAAGATTGG........................................................................................ | 20 | 2 | 4.00 | 4.00 | - | - | 1.00 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | 0.50 | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................AGGGGCGAGTGGAGTTTGGGA................................................................................................ | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................GTGGAGTTTGGGAAAGATTGGC....................................................................................... | 22 | 2 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................TGGAGTTTGGGAAAGATTG......................................................................................... | 19 | 2 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................TGGAGTTTGGGAAAGATTGGCATT.................................................................................... | 24 | 1 | 2.00 | 48.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................AGGGGCGAGTGGAGTTTGGTAAA.............................................................................................. | 23 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................AGTGGAGTTTGGGAAAGACTGG........................................................................................ | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................................................TGTTGCAGTTGGTTCGGCC..................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................................................TGTTGCAGTTGGTTCGG....................... | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................AGGGGCGAGTGGAGTTTGGGAAAGAC........................................................................................... | 26 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................TGGAGTTTGGGAAAGATTGGCATTA................................................................................... | 25 | 1 | 1.00 | 48.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................ATTCTTCCTCTTTGCTCCAGTA................................................ | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................AAAAACAGTCTTGAACAGC................................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................AGTGGAGTTTGGGAAAGATTGGA....................................................................................... | 23 | 2 | 1.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................CCAGTAAAATCAGAAAGTGAAAAAACAGTCTT........................................................................................................................ | 32 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - |
| ..................................................................................................................................................................................................................TGTTGCAGTTGGTTCGGT...................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................TGGAGTTTGGGAAAGATTGGCAGA.................................................................................... | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................................................................TTCCTGCAGTGAGCATGCA..... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................AAAAAACAGTCTTGAAGGAA................................................................................................................. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................................................................................................CAGTTGGTTCATTCCTGC................. | 18 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................AGGGGCGAGTGGAGTTTGGGC................................................................................................ | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................AGTGGAGTTTGGGAAAGATTGGT....................................................................................... | 23 | 2 | 0.50 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................GTTGCAGTTGGTTCATTCCTG.................. | 21 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................TGGAGTTTGGGAAAGATTGGCT...................................................................................... | 22 | 2 | 0.50 | 10.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................AGTGGAGTTTGGGAAAGAT........................................................................................... | 19 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................TTCATTCCTGCAGTGAGCA......... | 19 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - |
| ......................................................................................................................................................................................TCTTCCTCTTTGCTCCAG.................................................. | 18 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - |
| ..............................................................................................................................................TGGAGTTTGGGAAAGAT........................................................................................... | 17 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................AGACATAGAGCTTTGC....................................................................................................................................................... | 16 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 |
| TGGCTGCAGTGTCTGGCAGATTGGAAAAGGGATGATATCAGAAGGAGACAGACGGGTCAGAACATTCAGGAAAACTTCAAGTGAGACATAGAGCTTTGCCAGTAAAATCAGAAAGTGAAAAAACAGTCTTGAAAGGGGCGAGTGGAGTTTGGGAAAGATTGGCAGCCCAAGGTTTAAACCATTCTTCCTCTTTGCTCCAGGTGACCTCCTTGTTGCAGTTGGTTCATTCCTGCAGTGAGCAGTCTCCTCA ..............................................................................................................................................((((((..(((.((((.(((.((((...))))....))).)))).)))...))))))................................................... .........................................................................................................................................138...........................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR037938(GSM510476) 293Red. (cell line) | SRR207114(GSM721076) IP against AGO 1 & 2, RRP40 knockdown. (ago1/2 RRP40 cell line) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR038861(GSM458544) MM466. (cell line) | SRR033730(GSM497075) Lymphoblastic (ALL411). (B cell) | SRR033729(GSM497074) splenic MZL (Splenic414). (B cell) | SRR207116(GSM721078) Nuclear RNA. (cell line) | GSM1105748AGO1(GSM1105748) small RNA sequencing data. (ago1 hela) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | TAX577739(Rovira) total RNA. (breast) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | TAX577743(Rovira) total RNA. (breast) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR060982(GSM569186) Human centrocyte [09-001]. (cell line) | SRR038859(GSM458542) MM386. (cell line) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR326281(GSM769511) Dicer mRNA was knocked down using siDicer, cy. (cell line) | DRR000555(DRX000313) THP-1 whole cell RNA, no treatment. (cell line) | SRR038856(GSM458539) D11. (cell line) | SRR060983(GSM569187) Human pre-germinal center B cell [09-001]. (cell line) | SRR038858(GSM458541) MEL202. (cell line) | SRR040028(GSM532913) G026N. (cervix) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR033719(GSM497064) 6hr Activated B cell line (ABC158). (B cell) | SRR033723(GSM497068) L1236 cell line (L1236). (B cell) | SRR033717(GSM497062) Mentle Cell Lymphoma (MCL112). (B cell) | SRR189786 | SRR033716(GSM497061) Mentle Cell Lymphoma (MCL114). (B cell) | SRR330894(SRX091732) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033711(GSM497056) GCB DLBCL (GCB110). (B cell) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | DRR000562(DRX000320) Isolation of RNA following immunoprecipitatio. (ago3 cell line) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189787 | SRR033720(GSM497065) EBV activated B cell line (EBV159). (B cell) | SRR039193(GSM494812) HL60 cell line is derived from acute promyelo. (cell line) | DRR000558(DRX000316) THP-1 whole cell RNA, after 3 day treatment w. (cell line) | GSM1105749AGO2(GSM1105749) small RNA sequencing data. (ago2 hela) | SRR033732(GSM497077) bjab cell line (bjab103). (B cell) | SRR189782 | SRR060985(GSM569189) Human naive B cell [09-001]. (cell line) | TAX577453(Rovira) total RNA. (breast) | DRR000557(DRX000315) THP-1 whole cell RNA, after 3 day treatment w. (cell line) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR039637(GSM518474) THP1_total_sRNAs. (cell line) | SRR037933(GSM510471) 293cand4_rep2. (cell line) | GSM416733(GSM416733) HEK293. (cell line) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR033715(GSM497060) Mantle Cell Lymphoma (Mino122). (B cell) | SRR037931(GSM510469) 293GFP. (cell line) | SRR029126(GSM416755) 143B. (cell line) | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | DRR000561(DRX000319) Isolation of RNA following immunoprecipitatio. (ago2 cell line) | SRR037936(GSM510474) 293cand1. (cell line) | GSM450603(GSM450603) miRNA sequencing raw reads from post-mortem s. (brain) | SRR037934(GSM510472) 293cand4_rep3. (cell line) | GSM532883(GSM532883) G871N. (cervix) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................................................CTCCAGGTGACCTCCTAAGG.................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................................................TGCTCCAGGTGACCTCCCA....................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |