| (1) AGO1.ip | (2) AGO2.ip | (9) B-CELL | (2) BRAIN | (2) BREAST | (20) CELL-LINE | (5) CERVIX | (1) FIBROBLAST | (4) HEART | (1) HELA | (4) LIVER | (1) OTHER | (1) RRP40.ip | (9) SKIN | (1) TESTES | (1) UTERUS | (1) XRN.ip |
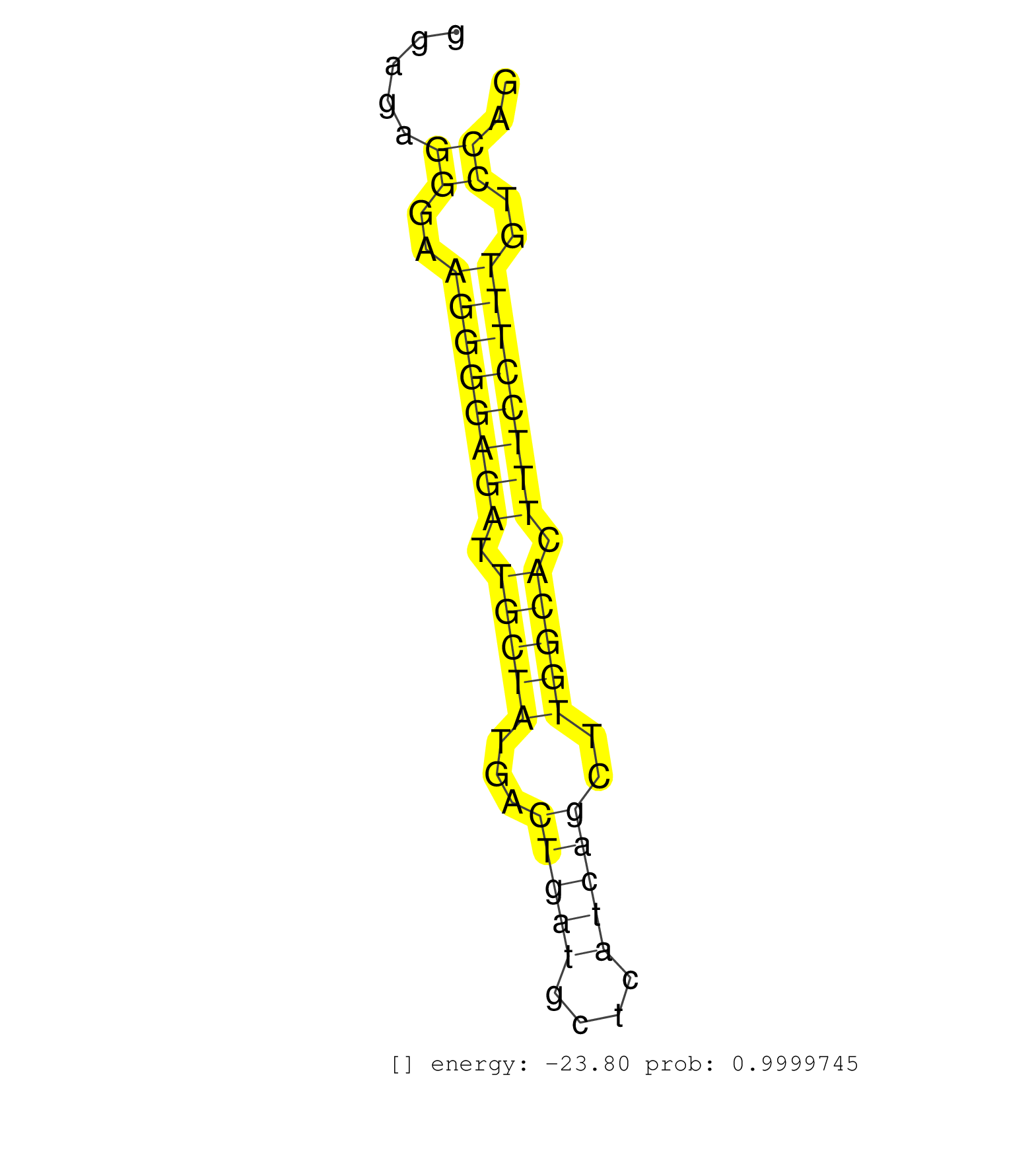
| AAGGTGCTGCTGGGAGGGGAGTCTGACAACCCAGCTAGCTGAAGCAGCAGGTGCCAGGCAGTAGGGCTGAGGCCTGGCTGGTTGGGAGAGGGAAGGGGAGATTGCTATGACTGATGCTCATCAGCTTGGCACTTTCCTTTGTCCAGAAGCGCCGGAGACCCACACTGGGGGTTCAGTTGGACGACAAACGCAAGGA .........................................................................................((..((((((((.(((((...(((((....)))))..))))).))))))))..)).................................................... ....................................................................................85...........................................................146................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR189782 | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | SRR038857(GSM458540) D20. (cell line) | SRR033715(GSM497060) Mantle Cell Lymphoma (Mino122). (B cell) | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | SRR060169(GSM565979) 5-8F_cytoplasm. (cell line) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR015360(GSM380325) Plasma B cells (PC137). (B cell) | SRR060986(GSM569190) Human memory B cell [09-001]. (cell line) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR553576(SRX182782) source: Testis. (testes) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR330895(SRX091733) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | DRR001486(DRX001040) Hela long cytoplasmic cell fraction, LNA(+). (hela) | SRR330891(SRX091729) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR040035(GSM532920) G001T. (cervix) | SRR038863(GSM458546) MM603. (cell line) | SRR039611(GSM531974) Human Normal Liver Tissue Sample 1. (liver) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039614(GSM531977) HBV-infected Liver Tissue. (liver) | SRR363674(GSM830251) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR015364(GSM380329) Plasma B cells (PC44). (B cell) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | SRR207110(GSM721072) Nuclear RNA. (cell line) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR039193(GSM494812) HL60 cell line is derived from acute promyelo. (cell line) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR189783 | SRR038856(GSM458539) D11. (cell line) | SRR037936(GSM510474) 293cand1. (cell line) | SRR040015(GSM532900) G623T. (cervix) | SRR040037(GSM532922) G243T. (cervix) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR040036(GSM532921) G243N. (cervix) | SRR191420(GSM715530) 122genomic small RNA (size selected RNA from . (breast) | GSM450607(GSM450607) miRNA sequencing raw reads from post-mortem s. (brain) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | GSM956925Ago2PAZ(GSM956925) cell line: HEK293 cell linepassages: 15-20ip . (ago2 cell line) | SRR040028(GSM532913) G026N. (cervix) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | DRR000556(DRX000314) THP-1 whole cell RNA, after 3 day treatment w. (cell line) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR015359(GSM380324) Germinal Center B cell (GC136). (B cell) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................GGGAAGGGGAGATTGCTATGACT.................................................................................... | 23 | 1 | 17.00 | 17.00 | 1.00 | - | - | - | 1.00 | - | - | 1.00 | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ........................................................................................AGGGAAGGGGAGATTGCTATGACT.................................................................................... | 24 | 1 | 5.00 | 5.00 | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ........................................................................................AGGGAAGGGGAGATTGCTAT........................................................................................ | 20 | 1 | 5.00 | 5.00 | 3.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................AGGGAAGGGGAGATTGCTATGA...................................................................................... | 22 | 1 | 4.00 | 4.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................GGGAAGGGGAGATTGCTATGAC..................................................................................... | 22 | 1 | 4.00 | 4.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................CTTGGCACTTTCCTTTGTCCAG.................................................. | 22 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................GGGAAGGGGAGATTGCTATG....................................................................................... | 20 | 1 | 3.00 | 3.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................GGGAAGGGGAGATTGCTATGACA.................................................................................... | 23 | 1 | 2.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................GGGAAGGGGAGATTGCTATGA...................................................................................... | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..............................................................................................................................................................CCCACACTGGGGGTTCAGTTGGACGACA.......... | 28 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................CTTGGCACTTTCCTTTGTCCAGT................................................. | 23 | 1 | 2.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................GCTAGCTGAAGCAGCAG.................................................................................................................................................. | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................AGGCAGTAGGGCTGAGGCCTGG....................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................CCCACACTGGGGGTTCAGTTGGACGACAAACGC..... | 33 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................CCCACACTGGGGGTTCAGTTGGA............... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................AGGGAAGGGGAGATTGCTATGAC..................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - |
| .......................TGACAACCCAGCTAGCTGAAGCAGCAGA................................................................................................................................................. | 28 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................CACACTGGGGGTTCAGTTGG................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................AGCGCCGGAGACCCAGGC............................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................AAGCGCCGGAGACCCACACTGGGGGC........................ | 26 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................CTTGGCACTTTCCTTTGTCCATT................................................. | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................................................................GGTTCAGTTGGACGACAA......... | 18 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................TCAGTTGGACGACAAACG...... | 18 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................CACTGGGGGTTCAGTTGGACGAC........... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................ACACTGGGGGTTCAGTTGGACGA............ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..........................................AGCAGCAGGTGCCAGGCAGAGGG................................................................................................................................... | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................................GGGGGTTCAGTTGGACGA............ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................GGGAAGGGGAGATTGCTAT........................................................................................ | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................CTGGGGGTTCAGTTGGACGACAAACGC..... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................CACTGGGGGTTCAGTGG................. | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | |
| ............................................CAGCAGGTGCCAGGCCATG..................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................AGGGAAGGGGAGATTGCTATGAGT.................................................................................... | 24 | 1 | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................CTGGGGGTTCAGTTGGAC.............. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................GGGAAGGGGAGATTGCTATGACAA................................................................................... | 24 | 1 | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................CACTGGGGGTTCAGTTGGACGACAAACGCAAGGA | 34 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................TTCAGTTGGACGACAAACTCTG... | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................CTTGGCACTTTCCTTTG....................................................... | 17 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................CCCACACTGGGGGTTCAGTTGGACGAC........... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................GTTCAGTTGGACGACAAACGCAAGGA | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| AAGGTGCTGCTGGGAGGGGAGTCTGACAACCCAGCTAGCTGAAGCAGCAGGTGCCAGGCAGTAGGGCTGAGGCCTGGCTGGTTGGGAGAGGGAAGGGGAGATTGCTATGACTGATGCTCATCAGCTTGGCACTTTCCTTTGTCCAGAAGCGCCGGAGACCCACACTGGGGGTTCAGTTGGACGACAAACGCAAGGA .........................................................................................((..((((((((.(((((...(((((....)))))..))))).))))))))..)).................................................... ....................................................................................85...........................................................146................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR189782 | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | SRR038857(GSM458540) D20. (cell line) | SRR033715(GSM497060) Mantle Cell Lymphoma (Mino122). (B cell) | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | SRR060169(GSM565979) 5-8F_cytoplasm. (cell line) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR015360(GSM380325) Plasma B cells (PC137). (B cell) | SRR060986(GSM569190) Human memory B cell [09-001]. (cell line) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR553576(SRX182782) source: Testis. (testes) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR330895(SRX091733) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | DRR001486(DRX001040) Hela long cytoplasmic cell fraction, LNA(+). (hela) | SRR330891(SRX091729) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR040035(GSM532920) G001T. (cervix) | SRR038863(GSM458546) MM603. (cell line) | SRR039611(GSM531974) Human Normal Liver Tissue Sample 1. (liver) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039614(GSM531977) HBV-infected Liver Tissue. (liver) | SRR363674(GSM830251) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR015364(GSM380329) Plasma B cells (PC44). (B cell) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | SRR207110(GSM721072) Nuclear RNA. (cell line) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR039193(GSM494812) HL60 cell line is derived from acute promyelo. (cell line) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR189783 | SRR038856(GSM458539) D11. (cell line) | SRR037936(GSM510474) 293cand1. (cell line) | SRR040015(GSM532900) G623T. (cervix) | SRR040037(GSM532922) G243T. (cervix) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR040036(GSM532921) G243N. (cervix) | SRR191420(GSM715530) 122genomic small RNA (size selected RNA from . (breast) | GSM450607(GSM450607) miRNA sequencing raw reads from post-mortem s. (brain) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | GSM956925Ago2PAZ(GSM956925) cell line: HEK293 cell linepassages: 15-20ip . (ago2 cell line) | SRR040028(GSM532913) G026N. (cervix) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | DRR000556(DRX000314) THP-1 whole cell RNA, after 3 day treatment w. (cell line) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR015359(GSM380324) Germinal Center B cell (GC136). (B cell) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................................GTTGGACGACAAACGCGGTT. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | |
| ...........GGGAGGGGAGTCTGAGTT....................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................AGCAGGTGCCAGGCAGTCAC................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................................TTGGACGACAAACGC..... | 15 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...........................AACCCAGCTAGCTGATGTA...................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................GGACGACAAACG...... | 12 | 7 | 0.29 | 0.29 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.29 |
| ..........TGGGAGGGGAGTCTGA.......................................................................................................................................................................... | 16 | 4 | 0.25 | 0.25 | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................CAGTTGGACGACA.......... | 13 | 8 | 0.12 | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |