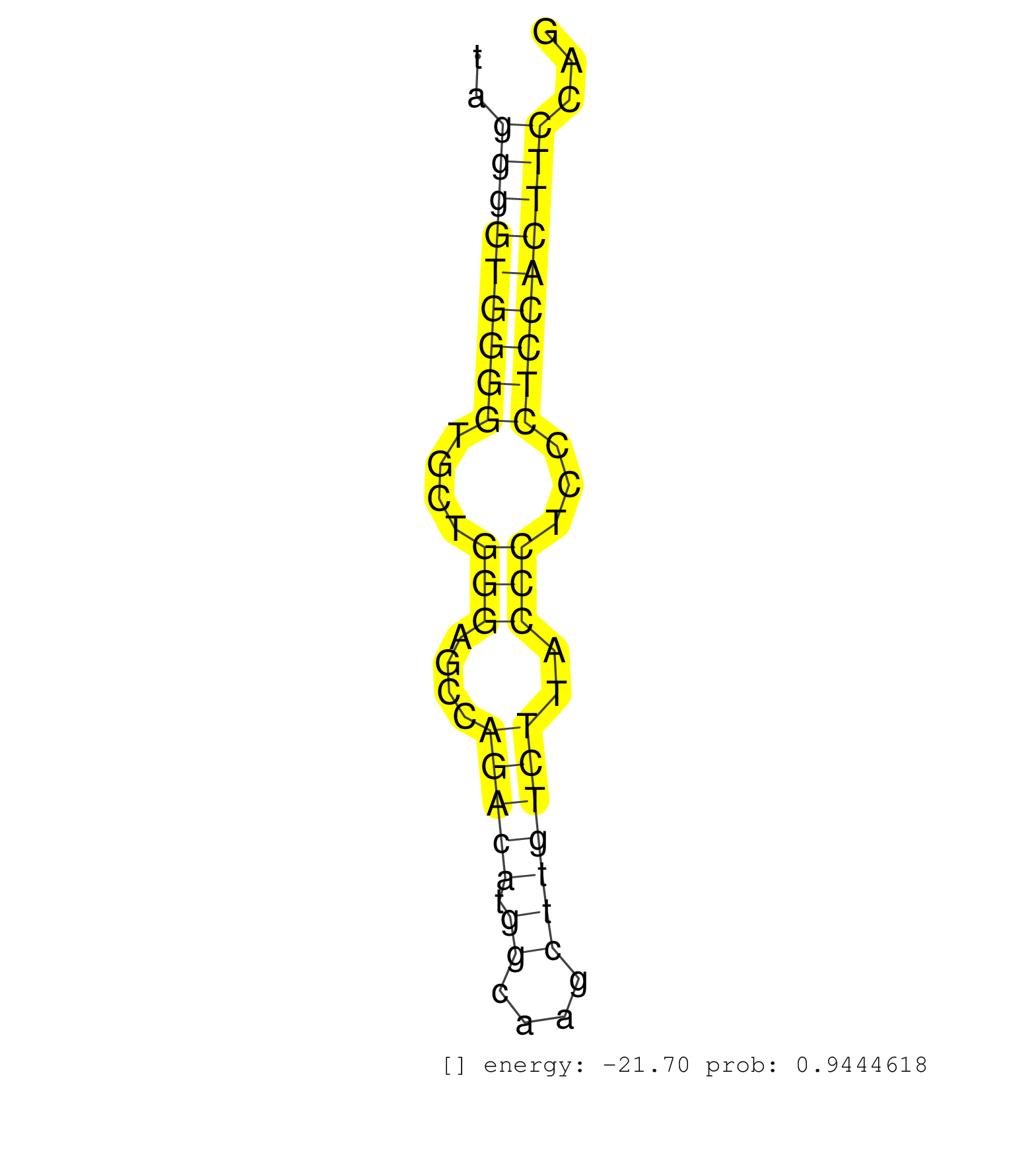
| (2) AGO1.ip | (3) AGO2.ip | (1) AGO3.ip | (2) B-CELL | (2) BRAIN | (6) BREAST | (11) CELL-LINE | (4) CERVIX | (1) FIBROBLAST | (3) HEART | (2) HELA | (2) LIVER | (1) OTHER | (18) SKIN | (1) XRN.ip |
| CACCAAGGCCACACTGCTGCGCCACATGCGGGAACGCCACTTCCGTCCAGGTGCGTCAGTAGCAGCCTACCTGCTTGGACTGCATGGGTAGGGGTGGGGTGCTGGGAGCCAGACATGGCAAGCTTGTCTTACCCTCCCTCCACTTCCAGTAGCAGCAGCCGCAGCAGCAGCTGGTAAAAAAGGACGTCTACGGAAGTGG ..........................................................................................(((((((((....(((....(((((.((....)))))))..)))...)))))))))..................................................... ........................................................................................89..........................................................149................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | SRR189782 | SRR207110(GSM721072) Nuclear RNA. (cell line) | SRR189784 | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | TAX577738(Rovira) total RNA. (breast) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | SRR037935(GSM510473) 293cand3. (cell line) | GSM1105748AGO1(GSM1105748) small RNA sequencing data. (ago1 hela) | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) | SRR040028(GSM532913) G026N. (cervix) | SRR330885(SRX091723) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR444040(SRX128888) Sample 1cDNABarcode: AF-PP-333: ACG CTC TTC C. (skin) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR330865(SRX091703) tissue: skin psoriatic involveddisease state:. (skin) | DRR000561(DRX000319) Isolation of RNA following immunoprecipitatio. (ago2 cell line) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR033719(GSM497064) 6hr Activated B cell line (ABC158). (B cell) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | TAX577580(Rovira) total RNA. (breast) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR191632(GSM715742) 78genomic small RNA (size selected RNA from t. (breast) | SRR039624(GSM531987) HBV(-) HCV(-) Adjacent Tissue Sample. (liver) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | SRR189785 | DRR000562(DRX000320) Isolation of RNA following immunoprecipitatio. (ago3 cell line) | SRR330888(SRX091726) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR040035(GSM532920) G001T. (cervix) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR343335 | SRR039635(GSM518472) THP1_nuc_sRNAs. (cell line) | SRR330870(SRX091708) tissue: skin psoriatic involveddisease state:. (skin) | SRR189787 | SRR330891(SRX091729) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR040030(GSM532915) G013N. (cervix) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191460(GSM715570) 33genomic small RNA (size selected RNA from t. (breast) | SRR191624(GSM715734) 31genomic small RNA (size selected RNA from t. (breast) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR095854(SRX039177) miRNA were isolated from FirstChoice Human Br. (brain) | SRR363674(GSM830251) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | GSM1105753INPUT(GSM1105753) small RNA sequencing data. (hela) | SRR033720(GSM497065) EBV activated B cell line (EBV159). (B cell) | SRR039193(GSM494812) HL60 cell line is derived from acute promyelo. (cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | GSM532882(GSM532882) G696T. (cervix) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................TCTTACCCTCCCTCCACTTCCAGT................................................. | 24 | 1 | 15.00 | 15.00 | 3.00 | 1.00 | - | - | 1.00 | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..............................................................................................................................TCTTACCCTCCCTCCACTTCCAG.................................................. | 23 | 1 | 13.00 | 13.00 | - | 6.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................GTGGGGTGCTGGGAGCCAGA...................................................................................... | 20 | 1 | 8.00 | 8.00 | - | - | 6.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................TCTTACCCTCCCTCCACTTCCA................................................... | 22 | 1 | 6.00 | 6.00 | - | - | - | - | - | - | - | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .............................................................................................GTGGGGTGCTGGGAGCCAGACA.................................................................................... | 22 | 1 | 6.00 | 6.00 | 2.00 | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................GTAGGGGTGGGGTGCTGGGAGCCAGA...................................................................................... | 26 | 1 | 5.00 | 5.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................TCTTACCCTCCCTCCACTTCC.................................................... | 21 | 1 | 4.00 | 4.00 | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................TTACCCTCCCTCCACTTCCAGT................................................. | 22 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ................................................................................................................................TTACCCTCCCTCCACTTCCAGA................................................. | 22 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................TCTTACCCTCCCTCCACTTC..................................................... | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................TTACCCTCCCTCCACTTCCAG.................................................. | 21 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................GTGGGGTGCTGGGAGCCAGAAG.................................................................................... | 22 | 1 | 2.00 | 8.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ................................................................................................................................TTACCCTCCCTCCACTTTCAG.................................................. | 21 | 2.00 | 0.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................TGGGGTGCTGGGAGCCAGA...................................................................................... | 19 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................AACGCCACTTCCGTCCAG..................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................AAAAAAGGACGTCTACGGAAGT.. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ........................................................................................................................................................CAGCAGCCGCAGCAGCAGCAAC......................... | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................CTCCCTCCACTTCCAGGG................................................ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | |
| ..........................................................................................................AGCCAGACATGGCAACGA........................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................TCCAGTAGCAGCAGCCCCAG................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................TCTTACCCTCCCTCCACTTCCAC.................................................. | 23 | 1 | 1.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................TCTTACCCTCCCTCCACTTCCAGC................................................. | 24 | 1 | 1.00 | 13.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................AAAAAAGGACGTCTACGGAAGTGG | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................TGGGGTGCTGGGAGCCAGACATAG................................................................................. | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................GCAGCAGCAGCTGGTAAAAAAGGACGTCTACGGA..... | 34 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................AAGGACGTCTACGGAAGT.. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................TCTTACCCTCCCTCCACTTCCAGA................................................. | 24 | 1 | 1.00 | 13.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................GGTGGGGTGCTGGGAGCCAG....................................................................................... | 20 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................AAAAGGACGTCTACGGAAGTG. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................TCTTACCCTCCCTCCACTTCCAT.................................................. | 23 | 1 | 1.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................CAGCAGCTGGTAAAAACTG................ | 19 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................................CAGCCGCAGCAGCAGTCGT......................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................................GCAGCTGGTAAAAAAGGACGTCTACGG...... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| .......................................................................TGCTTGGACTGCATGG................................................................................................................ | 16 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................GGGGTGGGGTGCTGGGA............................................................................................ | 17 | 9 | 0.11 | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 |
| CACCAAGGCCACACTGCTGCGCCACATGCGGGAACGCCACTTCCGTCCAGGTGCGTCAGTAGCAGCCTACCTGCTTGGACTGCATGGGTAGGGGTGGGGTGCTGGGAGCCAGACATGGCAAGCTTGTCTTACCCTCCCTCCACTTCCAGTAGCAGCAGCCGCAGCAGCAGCTGGTAAAAAAGGACGTCTACGGAAGTGG ..........................................................................................(((((((((....(((....(((((.((....)))))))..)))...)))))))))..................................................... ........................................................................................89..........................................................149................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | SRR189782 | SRR207110(GSM721072) Nuclear RNA. (cell line) | SRR189784 | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | TAX577738(Rovira) total RNA. (breast) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | SRR037935(GSM510473) 293cand3. (cell line) | GSM1105748AGO1(GSM1105748) small RNA sequencing data. (ago1 hela) | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) | SRR040028(GSM532913) G026N. (cervix) | SRR330885(SRX091723) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR444040(SRX128888) Sample 1cDNABarcode: AF-PP-333: ACG CTC TTC C. (skin) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR330865(SRX091703) tissue: skin psoriatic involveddisease state:. (skin) | DRR000561(DRX000319) Isolation of RNA following immunoprecipitatio. (ago2 cell line) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR033719(GSM497064) 6hr Activated B cell line (ABC158). (B cell) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | TAX577580(Rovira) total RNA. (breast) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR191632(GSM715742) 78genomic small RNA (size selected RNA from t. (breast) | SRR039624(GSM531987) HBV(-) HCV(-) Adjacent Tissue Sample. (liver) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | SRR189785 | DRR000562(DRX000320) Isolation of RNA following immunoprecipitatio. (ago3 cell line) | SRR330888(SRX091726) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR040035(GSM532920) G001T. (cervix) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR343335 | SRR039635(GSM518472) THP1_nuc_sRNAs. (cell line) | SRR330870(SRX091708) tissue: skin psoriatic involveddisease state:. (skin) | SRR189787 | SRR330891(SRX091729) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR040030(GSM532915) G013N. (cervix) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191460(GSM715570) 33genomic small RNA (size selected RNA from t. (breast) | SRR191624(GSM715734) 31genomic small RNA (size selected RNA from t. (breast) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR095854(SRX039177) miRNA were isolated from FirstChoice Human Br. (brain) | SRR363674(GSM830251) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | GSM1105753INPUT(GSM1105753) small RNA sequencing data. (hela) | SRR033720(GSM497065) EBV activated B cell line (EBV159). (B cell) | SRR039193(GSM494812) HL60 cell line is derived from acute promyelo. (cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | GSM532882(GSM532882) G696T. (cervix) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................................CAGCAGCCGCAGCAGCAGAAC.......................... | 21 | 2.00 | 0.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................CCCTCCCTCCACTTCCAAGTG............................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................CTCCCTCCACTTCCAGTAGTG............................................. | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................AGGACGTCTACG....... | 12 | 8 | 0.12 | 0.12 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.12 | - |