| (2) AGO1.ip | (1) AGO1.ip OTHER.mut | (5) AGO2.ip | (1) AGO3.ip | (2) B-CELL | (6) BRAIN | (12) BREAST | (32) CELL-LINE | (1) FIBROBLAST | (4) HEART | (2) HELA | (1) KIDNEY | (1) LIVER | (3) OTHER | (19) SKIN | (1) UTERUS | (1) XRN.ip |
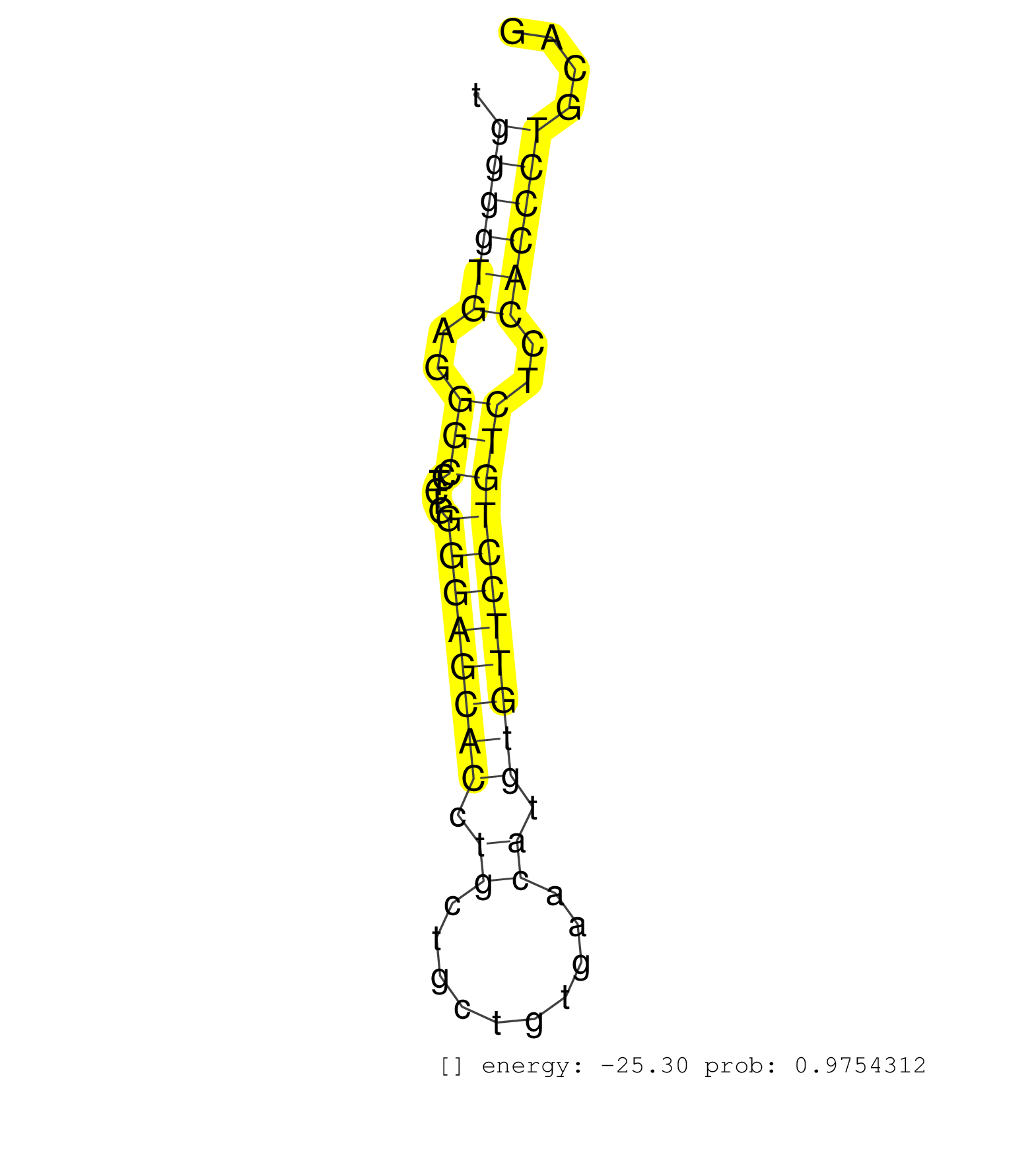
| GCTTCATCCCTGGACAAGGCGCGCGGCCACCACAGCAAGGAAATGGCCCTTTCAGTGCTAGGAAGTGTCGCGTTCGGCAGGAGACTGGGATGGAGCGGGGCCTGGGGGTGAAGAGGGTCCTCATGAGCCGCGTGTGTGGGGTGAGGGCCTCTGGGGAGCACCTGCTGCTGTGAACATGTGTTCCTGTCTCCACCCTGCAGTGCGGCATCGTCCTGGGGGCGCTGCTCTTGGTCTTCTGCTCATGGATGAC .........................................................................................................................................((((((..(((.....((((((((.((..........)).)))))))))))..))))))...................................................... ........................................................................................................................................137............................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | SRR207114(GSM721076) IP against AGO 1 & 2, RRP40 knockdown. (ago1/2 RRP40 cell line) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR189782 | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR387909(GSM843861) specific-host: Homo sapienshost cell line: Su. (cell line) | DRR000557(DRX000315) THP-1 whole cell RNA, after 3 day treatment w. (cell line) | DRR000555(DRX000313) THP-1 whole cell RNA, no treatment. (cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR207110(GSM721072) Nuclear RNA. (cell line) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR553575(SRX182781) source: Kidney. (Kidney) | SRR037933(GSM510471) 293cand4_rep2. (cell line) | SRR191592(GSM715702) 59genomic small RNA (size selected RNA from t. (breast) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | DRR000560(DRX000318) Isolation of RNA following immunoprecipitatio. (ago1 cell line) | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) | TAX577746(Rovira) total RNA. (breast) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | GSM450604(GSM450604) miRNA sequencing raw reads from post-mortem s. (brain) | DRR000561(DRX000319) Isolation of RNA following immunoprecipitatio. (ago2 cell line) | SRR037935(GSM510473) 293cand3. (cell line) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | DRR001489(DRX001043) Hela short nuclear cell fraction, control. (hela) | GSM416733(GSM416733) HEK293. (cell line) | SRR191431(GSM715541) 168genomic small RNA (size selected RNA from . (breast) | SRR189786 | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR330894(SRX091732) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR038859(GSM458542) MM386. (cell line) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR191540(GSM715650) 62genomic small RNA (size selected RNA from t. (breast) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330903(SRX091741) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR060982(GSM569186) Human centrocyte [09-001]. (cell line) | SRR060983(GSM569187) Human pre-germinal center B cell [09-001]. (cell line) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | GSM956925Ago2(GSM956925) cell line: HEK293 cell linepassages: 15-20ip . (ago2 cell line) | SRR330883(SRX091721) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577590(Rovira) total RNA. (breast) | SRR029128(GSM416757) H520. (cell line) | TAX577745(Rovira) total RNA. (breast) | SRR037941(GSM510479) 293DroshaTN. (cell line) | SRR330860(SRX091698) tissue: skin psoriatic involveddisease state:. (skin) | TAX577738(Rovira) total RNA. (breast) | SRR033729(GSM497074) splenic MZL (Splenic414). (B cell) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR330869(SRX091707) tissue: skin psoriatic involveddisease state:. (skin) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | DRR000556(DRX000314) THP-1 whole cell RNA, after 3 day treatment w. (cell line) | SRR444040(SRX128888) Sample 1cDNABarcode: AF-PP-333: ACG CTC TTC C. (skin) | SRR015448(SRR015448) cytoplasmic small RNAs. (breast) | SRR037936(GSM510474) 293cand1. (cell line) | SRR037934(GSM510472) 293cand4_rep3. (cell line) | GSM450603(GSM450603) miRNA sequencing raw reads from post-mortem s. (brain) | SRR037943(GSM510481) 293DcrTN. (cell line) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR330913(SRX091751) tissue: normal skindisease state: normal. (skin) | SRR363676(GSM830253) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | TAX577744(Rovira) total RNA. (breast) | SRR037931(GSM510469) 293GFP. (cell line) | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | GSM1105749AGO2(GSM1105749) small RNA sequencing data. (ago2 hela) | TAX577742(Rovira) total RNA. (breast) | SRR039191(GSM494810) PBMCs were isolated by ficoll gradient from t. (blood) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | DRR000562(DRX000320) Isolation of RNA following immunoprecipitatio. (ago3 cell line) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | SRR191528(GSM715638) 130genomic small RNA (size selected RNA from . (breast) | SRR326282(GSM769512) Dicer mRNA was knocked down using siDicer, to. (cell line) | SRR330923(SRX091761) tissue: normal skindisease state: normal. (skin) | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | SRR037932(GSM510470) 293cand4_rep1. (cell line) | SRR330895(SRX091733) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR189783 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................................................GTTCCTGTCTCCACCCTGCAGA................................................. | 22 | 1 | 34.00 | 4.00 | 10.00 | - | - | 2.00 | 2.00 | - | - | - | 1.00 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ....................................................................................................................................................................................TTCCTGTCTCCACCCTGCAGA................................................. | 21 | 1 | 17.00 | 4.00 | - | - | 1.00 | - | 2.00 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TGTTCCTGTCTCCACCCTGCAGT................................................. | 23 | 1 | 8.00 | 8.00 | - | 8.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................GTTCCTGTCTCCACCCTGCAGT................................................. | 22 | 1 | 7.00 | 7.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - |
| ....................................................................................................................................................................................TTCCTGTCTCCACCCTGCAGAA................................................ | 22 | 1 | 5.00 | 4.00 | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TTCCTGTCTCCACCCTGCAGT................................................. | 21 | 1 | 5.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ...................................................................................................................................................................................GTTCCTGTCTCCACCCTGCAGAA................................................ | 23 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................GTTCCTGTCTCCACCCTGCAG.................................................. | 21 | 1 | 4.00 | 4.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TTCCTGTCTCCACCCTGCAGAAT............................................... | 23 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TTCCTGTCTCCACCCTGCAG.................................................. | 20 | 1 | 4.00 | 4.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................GTTCCTGTCTCCACCCTGCAGC................................................. | 22 | 1 | 4.00 | 4.00 | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................GTTCCTGTCTCCACCCTGCA................................................... | 20 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TGTTCCTGTCTCCACCCTGCAGA................................................. | 23 | 3.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................TTCCTGTCTCCACCCTGCAGAC................................................ | 22 | 1 | 3.00 | 4.00 | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................................TTCTGCTCATGGATGAC | 17 | 3 | 2.67 | 2.67 | - | - | - | - | - | - | - | - | - | - | - | 2.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................GTTCCTGTCTCCACCCTGCAGATC............................................... | 24 | 1 | 2.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TGTGTTCCTGTCTCCACCCTGC.................................................... | 22 | 1 | 2.00 | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TTCCTGTCTCCACCCTGCAGTT................................................ | 22 | 1 | 2.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TTCCTGTCTCCACCCTGCAGAAA............................................... | 23 | 1 | 2.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................GTTCCTGTCTCCACCCTGCCGA................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................TGCTCTTGGTCTTCTGCTCATG...... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ........................................................................TTCGGCAGGAGACTGTC................................................................................................................................................................. | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................................TGTGTTCCTGTCTCCACCCAAA.................................................... | 22 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................GTTCCTGTCTCCACCCTGCAGAT................................................ | 23 | 1 | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................AGTGCGGCATCGTCCTGGGG................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................GTTCCTGTCTCCACCCTG..................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................GTTCCTGTCTCCACCCTGCAGAAT............................................... | 24 | 1 | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TTCCTGTCTCCACCCTGCAGAGT............................................... | 23 | 1 | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................CCTGCTGCTGTGAACACTGG...................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................................................................................CTCTTGGTCTTCTGCTTG........ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................GGCCTGGGGGTGAAGAGATGC................................................................................................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................TGGAGCGGGGCCTGGGGCCGC........................................................................................................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................TTCCTGTCTCCACCCTGCAGAATC.............................................. | 24 | 1 | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TTCCTGTCTCCACCCTGCCGC................................................. | 21 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................TGAGGGCCTCTGGGGAGCAC......................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................GTTCCTGTCTCCACCCTGCAAAA................................................ | 23 | 1 | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TTCCTGTCTCCACCCTGCAGC................................................. | 21 | 1 | 1.00 | 4.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TTCCTGTCTCCACCCTGCAGAGA............................................... | 23 | 1 | 1.00 | 4.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................GTTCCTGTCTCCACCCTGC.................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................TGGGATGGAGCGGGGTAG................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................TGGGGGTGAAGAGGGTCCTCA............................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TGTTCCTGTCTCCACCCTGCAGATT............................................... | 25 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................GTTCCTGTCTCCACCCTGCAA.................................................. | 21 | 1 | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................TGTGTGGGGTGAGGGTAA.................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | |
| ....................................................................................................................................................................................TTCCTGTCTCCACCCTGCAGTTA............................................... | 23 | 1 | 1.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TTCCTGTCTCCACCCTGCAGAT................................................ | 22 | 1 | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TTCCTGTCTCCACCCTGCAGATA............................................... | 23 | 1 | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................GTTCCTGTCTCCACCCTGCATTA................................................ | 23 | 1 | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................GTGCGGCATCGTCCTGG.................................. | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TTCCTGTCTCCACCCTGCAGAGC............................................... | 23 | 1 | 1.00 | 4.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................TGTCGCGTTCGG............................................................................................................................................................................. | 12 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................GAGCGGGGCCTGGGGGTG............................................................................................................................................ | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................CAAGGAAATGGCCCTTT...................................................................................................................................................................................................... | 17 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - |
| ......................................................................................................................................................................................................AGTGCGGCATCG........................................ | 12 | 9 | 0.11 | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 |
| GCTTCATCCCTGGACAAGGCGCGCGGCCACCACAGCAAGGAAATGGCCCTTTCAGTGCTAGGAAGTGTCGCGTTCGGCAGGAGACTGGGATGGAGCGGGGCCTGGGGGTGAAGAGGGTCCTCATGAGCCGCGTGTGTGGGGTGAGGGCCTCTGGGGAGCACCTGCTGCTGTGAACATGTGTTCCTGTCTCCACCCTGCAGTGCGGCATCGTCCTGGGGGCGCTGCTCTTGGTCTTCTGCTCATGGATGAC .........................................................................................................................................((((((..(((.....((((((((.((..........)).)))))))))))..))))))...................................................... ........................................................................................................................................137............................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | SRR207114(GSM721076) IP against AGO 1 & 2, RRP40 knockdown. (ago1/2 RRP40 cell line) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR189782 | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR387909(GSM843861) specific-host: Homo sapienshost cell line: Su. (cell line) | DRR000557(DRX000315) THP-1 whole cell RNA, after 3 day treatment w. (cell line) | DRR000555(DRX000313) THP-1 whole cell RNA, no treatment. (cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR207110(GSM721072) Nuclear RNA. (cell line) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR553575(SRX182781) source: Kidney. (Kidney) | SRR037933(GSM510471) 293cand4_rep2. (cell line) | SRR191592(GSM715702) 59genomic small RNA (size selected RNA from t. (breast) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | DRR000560(DRX000318) Isolation of RNA following immunoprecipitatio. (ago1 cell line) | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) | TAX577746(Rovira) total RNA. (breast) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | GSM450604(GSM450604) miRNA sequencing raw reads from post-mortem s. (brain) | DRR000561(DRX000319) Isolation of RNA following immunoprecipitatio. (ago2 cell line) | SRR037935(GSM510473) 293cand3. (cell line) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | DRR001489(DRX001043) Hela short nuclear cell fraction, control. (hela) | GSM416733(GSM416733) HEK293. (cell line) | SRR191431(GSM715541) 168genomic small RNA (size selected RNA from . (breast) | SRR189786 | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR330894(SRX091732) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR038859(GSM458542) MM386. (cell line) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR191540(GSM715650) 62genomic small RNA (size selected RNA from t. (breast) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330903(SRX091741) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR060982(GSM569186) Human centrocyte [09-001]. (cell line) | SRR060983(GSM569187) Human pre-germinal center B cell [09-001]. (cell line) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | GSM956925Ago2(GSM956925) cell line: HEK293 cell linepassages: 15-20ip . (ago2 cell line) | SRR330883(SRX091721) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577590(Rovira) total RNA. (breast) | SRR029128(GSM416757) H520. (cell line) | TAX577745(Rovira) total RNA. (breast) | SRR037941(GSM510479) 293DroshaTN. (cell line) | SRR330860(SRX091698) tissue: skin psoriatic involveddisease state:. (skin) | TAX577738(Rovira) total RNA. (breast) | SRR033729(GSM497074) splenic MZL (Splenic414). (B cell) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR330869(SRX091707) tissue: skin psoriatic involveddisease state:. (skin) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | DRR000556(DRX000314) THP-1 whole cell RNA, after 3 day treatment w. (cell line) | SRR444040(SRX128888) Sample 1cDNABarcode: AF-PP-333: ACG CTC TTC C. (skin) | SRR015448(SRR015448) cytoplasmic small RNAs. (breast) | SRR037936(GSM510474) 293cand1. (cell line) | SRR037934(GSM510472) 293cand4_rep3. (cell line) | GSM450603(GSM450603) miRNA sequencing raw reads from post-mortem s. (brain) | SRR037943(GSM510481) 293DcrTN. (cell line) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR330913(SRX091751) tissue: normal skindisease state: normal. (skin) | SRR363676(GSM830253) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | TAX577744(Rovira) total RNA. (breast) | SRR037931(GSM510469) 293GFP. (cell line) | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | GSM1105749AGO2(GSM1105749) small RNA sequencing data. (ago2 hela) | TAX577742(Rovira) total RNA. (breast) | SRR039191(GSM494810) PBMCs were isolated by ficoll gradient from t. (blood) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | DRR000562(DRX000320) Isolation of RNA following immunoprecipitatio. (ago3 cell line) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | SRR191528(GSM715638) 130genomic small RNA (size selected RNA from . (breast) | SRR326282(GSM769512) Dicer mRNA was knocked down using siDicer, to. (cell line) | SRR330923(SRX091761) tissue: normal skindisease state: normal. (skin) | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | SRR037932(GSM510470) 293cand4_rep1. (cell line) | SRR330895(SRX091733) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR189783 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................CGCGGCCACCACAGCTGG................................................................................................................................................................................................................... | 18 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................CGCGTTCGGCAGGAGCCA.................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................AAGTGTCGCGTTCG.............................................................................................................................................................................. | 14 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................GAGCGGGGCCTGGGGGCC............................................................................................................................................ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................GCGGGGCCTGGGGGTGATCC........................................................................................................................................ | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................CGCGGCCACCACAGCTGGG.................................................................................................................................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................................................................................GCGCTGCTCTTGGTCTTGA............. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................................................................................................................GGTCTTCTGCTCATGG..... | 16 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - |