| (1) AGO1.ip | (2) AGO2.ip | (1) AGO3.ip | (4) B-CELL | (2) BRAIN | (10) BREAST | (22) CELL-LINE | (4) CERVIX | (1) FIBROBLAST | (4) HEART | (6) HELA | (2) LIVER | (2) OTHER | (8) SKIN | (1) TESTES | (1) XRN.ip |
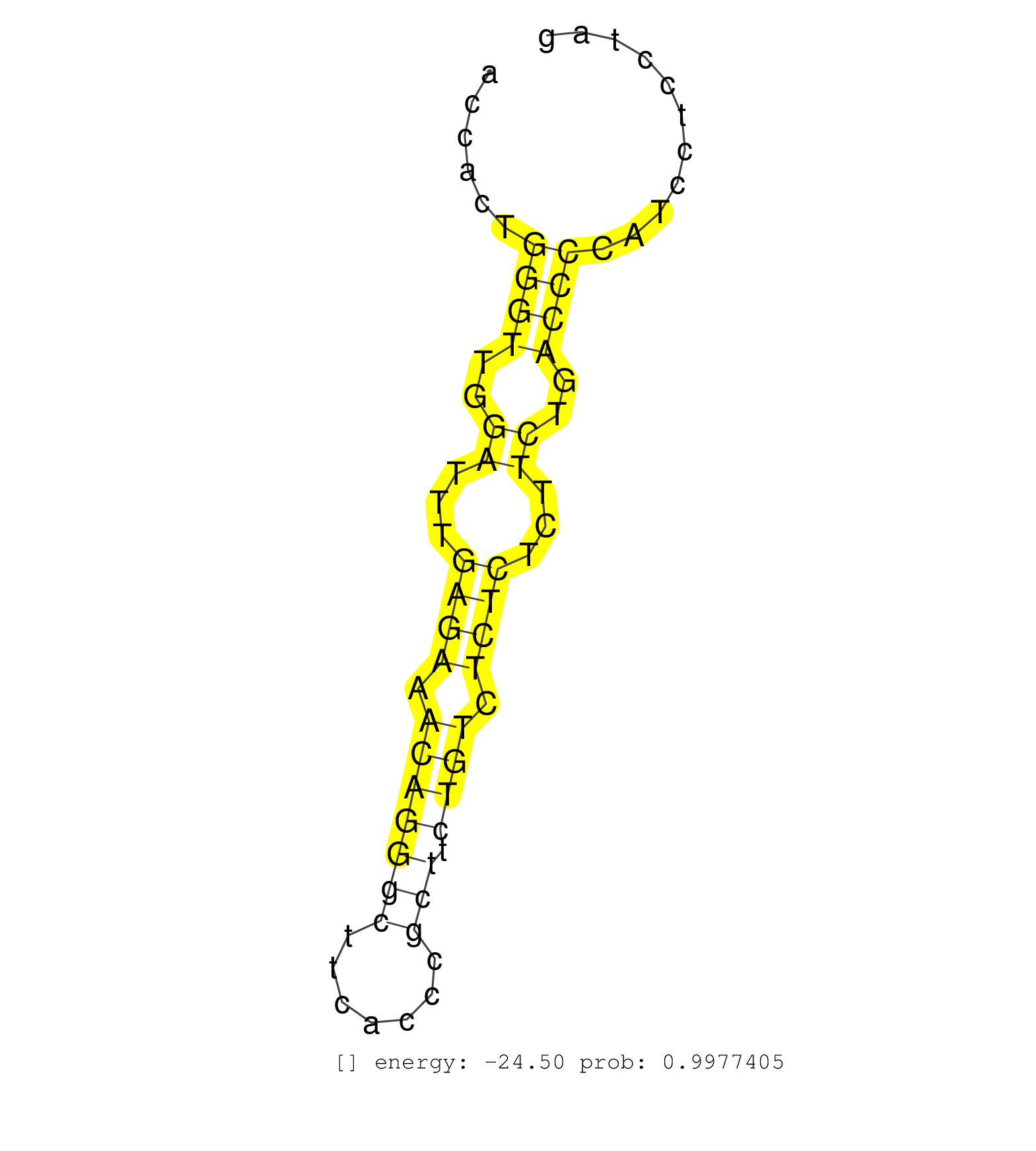
| CCTTTCCTCCTCTCTTTCCCTGGTCCTCTCCTGGTCTCCCATCTGTGGCACAGACTTCACCTCCCTTTCCTCTCACCTTGTGCTCCTGAAGGCCTCTGCCATCGCTTGGACCCCCTCGCCTCGCTGAGGACCACTGGGTTGGATTTGAGAAACAGGGCTTCACCCGCTTCTGTCTCTCTCTTCTGACCCCATCCTCCTAGGTGCGAAAGGGTTTCTGGAAACAGCCTCCCCTCATCTGGGATGTCAATCC .......................................................................................................................................((((..((...((((.(((((((.......))).)))).))))...))..))))............................................................. .................................................................................................................................130...................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR387909(GSM843861) specific-host: Homo sapienshost cell line: Su. (cell line) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR189782 | GSM339996(GSM339996) hues6Neuron. (cell line) | SRR207116(GSM721078) Nuclear RNA. (cell line) | GSM1105753INPUT(GSM1105753) small RNA sequencing data. (hela) | DRR001489(DRX001043) Hela short nuclear cell fraction, control. (hela) | SRR553576(SRX182782) source: Testis. (testes) | SRR189784 | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR326282(GSM769512) Dicer mRNA was knocked down using siDicer, to. (cell line) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR038861(GSM458544) MM466. (cell line) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR326281(GSM769511) Dicer mRNA was knocked down using siDicer, cy. (cell line) | DRR000557(DRX000315) THP-1 whole cell RNA, after 3 day treatment w. (cell line) | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR191456(GSM715566) 182genomic small RNA (size selected RNA from . (breast) | SRR040035(GSM532920) G001T. (cervix) | SRR038863(GSM458546) MM603. (cell line) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR033715(GSM497060) Mantle Cell Lymphoma (Mino122). (B cell) | SRR191468(GSM715578) 116genomic small RNA (size selected RNA from . (breast) | DRR000560(DRX000318) Isolation of RNA following immunoprecipitatio. (ago1 cell line) | SRR033723(GSM497068) L1236 cell line (L1236). (B cell) | SRR343334 | SRR033714(GSM497059) Burkitt Lymphoma (BL134). (B cell) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR189785 | GSM532876(GSM532876) G547T. (cervix) | SRR191450(GSM715560) 145genomic small RNA (size selected RNA from . (breast) | SRR343335 | SRR029132(GSM416761) MB-MDA231. (cell line) | SRR330904(SRX091742) tissue: normal skindisease state: normal. (skin) | SRR040011(GSM532896) G529T. (cervix) | SRR330898(SRX091736) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191464(GSM715574) 112genomic small RNA (size selected RNA from . (breast) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | SRR038856(GSM458539) D11. (cell line) | SRR038857(GSM458540) D20. (cell line) | SRR191590(GSM715700) 48genomic small RNA (size selected RNA from t. (breast) | TAX577579(Rovira) total RNA. (breast) | SRR038858(GSM458541) MEL202. (cell line) | SRR038852(GSM458535) QF1160MB. (cell line) | SRR040037(GSM532922) G243T. (cervix) | DRR001483(DRX001037) Hela long cytoplasmic cell fraction, control. (hela) | DRR001485(DRX001039) Hela long total cell fraction, LNA(+). (hela) | SRR330914(SRX091752) tissue: normal skindisease state: normal. (skin) | SRR553574(SRX182780) source: Heart. (Heart) | TAX577738(Rovira) total RNA. (breast) | SRR029124(GSM416753) HeLa. (hela) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR330911(SRX091749) tissue: normal skindisease state: normal. (skin) | SRR191605(GSM715715) 76genomic small RNA (size selected RNA from t. (breast) | SRR363673(GSM830250) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR037937(GSM510475) 293cand2. (cell line) | SRR191434(GSM715544) 171genomic small RNA (size selected RNA from . (breast) | SRR189783 | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | GSM1105750AGO3(GSM1105750) small RNA sequencing data. (ago3 hela) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | DRR000558(DRX000316) THP-1 whole cell RNA, after 3 day treatment w. (cell line) | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) | SRR330885(SRX091723) tissue: skin psoriatic uninvolveddisease stat. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................TGGGTTGGATTTGAGAAACAGG.............................................................................................. | 22 | 1 | 37.00 | 37.00 | 18.00 | 7.00 | 1.00 | - | 1.00 | - | - | - | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ....................................................................................................................................ACTGGGTTGGATTTGAGAAAC................................................................................................. | 21 | 1 | 28.00 | 28.00 | 22.00 | 1.00 | - | - | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................CTGGGTTGGATTTGAGAAACAGG.............................................................................................. | 23 | 1 | 23.00 | 23.00 | 2.00 | - | - | 10.00 | 3.00 | - | - | - | - | 1.00 | - | - | 2.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................ACTGGGTTGGATTTGAGAAACAG............................................................................................... | 23 | 1 | 19.00 | 19.00 | 11.00 | 2.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................ACTGGGTTGGATTTGAGAAACAGG.............................................................................................. | 24 | 1 | 15.00 | 15.00 | 7.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | 2.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................TGGGTTGGATTTGAGAAACAG............................................................................................... | 21 | 1 | 12.00 | 12.00 | 2.00 | 3.00 | 2.00 | - | 1.00 | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ....................................................................................................................................ACTGGGTTGGATTTGAGAAACA................................................................................................ | 22 | 1 | 11.00 | 11.00 | 4.00 | - | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................TGTCTCTCTCTTCTGACCCCAT.......................................................... | 22 | 1 | 7.00 | 7.00 | - | - | - | - | - | - | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................CTGGGTTGGATTTGAGAAACAG............................................................................................... | 22 | 1 | 5.00 | 5.00 | 1.00 | 1.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................TGGGTTGGATTTGAGAAACAGGA............................................................................................. | 23 | 1 | 5.00 | 37.00 | - | - | 3.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................ACTGGGTTGGATTTGAGAAA.................................................................................................. | 20 | 1 | 5.00 | 5.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................ACTGGGTTGGATTTGAGA.................................................................................................... | 18 | 1 | 4.00 | 4.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................ACTGGGTTGGATTTGAGAAACAA............................................................................................... | 23 | 1 | 3.00 | 11.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................CTGGGTTGGATTTGAGAAACAGGT............................................................................................. | 24 | 1 | 3.00 | 23.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - |
| ............................................................................................................................TGAGGACCACTGGGTTGGATTTGAG..................................................................................................... | 25 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................CTTCTGACCCCATCCTCCACCG................................................. | 22 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................CTGGGTTGGATTTGAGAAACA................................................................................................ | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................TGAGGACCACTGGGTTGGATTT........................................................................................................ | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................GAGGACCACTGGGTTGGATTTGAGAAAC................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................GGGTTGGATTTGAGAAACAGGGTT........................................................................................... | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................CTGGGTTGGATTTGAGAAA.................................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................CCCTCGCCTCGCTGAGGACCACTGGGTTGGATTTGAGAAACAGGGCTTCACCCGCTTCTGTCTCTCTCTTCTGACCCCATCCTCCTAGA................................................. | 89 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................CTGGGTTGGATTTGAGAACCAG............................................................................................... | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................................................................................................TGGAAACAGCCTCCCCTCTCCG............. | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................TGAGGACCACTGGGTTGGATTTGAGAAACAGGG............................................................................................. | 33 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TCTTCTGACCCCATCCTCCTAGA................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................ACTGGGTTGGATTTGAGAA................................................................................................... | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................CTGGTCTCCCATCTGAAC.......................................................................................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................TCTTCTGACCCCATCCTCCTAGGTGCA............................................. | 27 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................TGGGTTGGATTTGAGAAACAGA.............................................................................................. | 22 | 1 | 1.00 | 12.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TCTTCTGACCCCATCCTCCTAGAA................................................ | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................CTGGGTTGGATTTGAGAAACAGA.............................................................................................. | 23 | 1 | 1.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................CTGGGTTGGATTTGAGAAG.................................................................................................. | 19 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................TGAGGACCACTGGGTTGGGTTT........................................................................................................ | 22 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................ACTGGGTTGGATTTGAGAAACAGGC............................................................................................. | 25 | 1 | 1.00 | 15.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................GGATTTGAGAAACAGGGCTT.......................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| .......................................................................................................................................................................................................GGTGCGAAAGGGTTTCTGGAAACAGCC........................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................TGGGTTGGATTTGAGAAGCAG............................................................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................................TCCTAGGTGCGAAAGA........................................ | 16 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................TCTTCTGACCCCATCCTCCTAG.................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................CTGGGTTGGATTTGAGAAAC................................................................................................. | 20 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................GGGTTGGATTTGAGAAACAGG.............................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................TGAGGACCACTGGGTTGGATTTGAGAAACAG............................................................................................... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ............................................................................................................................TGAGGACCACTGGGTTGGATTTGAGAAAC................................................................................................. | 29 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................CTTGTGCTCCTGAAGGG............................................................................................................................................................. | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................ACTGGGTTGGATTTGAGAAACAGT.............................................................................................. | 24 | 1 | 1.00 | 19.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................ACTGGGTTGGATTTGAGAAACAGGGCTTCACC...................................................................................... | 32 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................GGTTTCTGGAAACAGCCTCCCC................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................TGGGTTGGATTTGAGAAAC................................................................................................. | 19 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................TGGGTTGGATTTGAGAAACAGGT............................................................................................. | 23 | 1 | 1.00 | 37.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................TTGGATTTGAGAAACAGGGCTTCA........................................................................................ | 24 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................GACCCCCTCGCCTCGGCGG........................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................................................................................CTGGAAACAGCCTCCCCTC................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................TCTCTCTCTTCTGACCCCATCCTCC..................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................TGGGTTGGATTTGAGAAAGA................................................................................................ | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................TGGGTTGGATTTGAGAAACAGGAA............................................................................................ | 24 | 1 | 1.00 | 37.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................GGGTTGGATTTGAGAAACAGGGCTTCA........................................................................................ | 27 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CCTTTCCTCCTCTCTTTCCCTGGTCCTCTCCTGGTCTCCCATCTGTGGCACAGACTTCACCTCCCTTTCCTCTCACCTTGTGCTCCTGAAGGCCTCTGCCATCGCTTGGACCCCCTCGCCTCGCTGAGGACCACTGGGTTGGATTTGAGAAACAGGGCTTCACCCGCTTCTGTCTCTCTCTTCTGACCCCATCCTCCTAGGTGCGAAAGGGTTTCTGGAAACAGCCTCCCCTCATCTGGGATGTCAATCC .......................................................................................................................................((((..((...((((.(((((((.......))).)))).))))...))..))))............................................................. .................................................................................................................................130...................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR387909(GSM843861) specific-host: Homo sapienshost cell line: Su. (cell line) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR189782 | GSM339996(GSM339996) hues6Neuron. (cell line) | SRR207116(GSM721078) Nuclear RNA. (cell line) | GSM1105753INPUT(GSM1105753) small RNA sequencing data. (hela) | DRR001489(DRX001043) Hela short nuclear cell fraction, control. (hela) | SRR553576(SRX182782) source: Testis. (testes) | SRR189784 | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR326282(GSM769512) Dicer mRNA was knocked down using siDicer, to. (cell line) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR038861(GSM458544) MM466. (cell line) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR326281(GSM769511) Dicer mRNA was knocked down using siDicer, cy. (cell line) | DRR000557(DRX000315) THP-1 whole cell RNA, after 3 day treatment w. (cell line) | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR191456(GSM715566) 182genomic small RNA (size selected RNA from . (breast) | SRR040035(GSM532920) G001T. (cervix) | SRR038863(GSM458546) MM603. (cell line) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR033715(GSM497060) Mantle Cell Lymphoma (Mino122). (B cell) | SRR191468(GSM715578) 116genomic small RNA (size selected RNA from . (breast) | DRR000560(DRX000318) Isolation of RNA following immunoprecipitatio. (ago1 cell line) | SRR033723(GSM497068) L1236 cell line (L1236). (B cell) | SRR343334 | SRR033714(GSM497059) Burkitt Lymphoma (BL134). (B cell) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR189785 | GSM532876(GSM532876) G547T. (cervix) | SRR191450(GSM715560) 145genomic small RNA (size selected RNA from . (breast) | SRR343335 | SRR029132(GSM416761) MB-MDA231. (cell line) | SRR330904(SRX091742) tissue: normal skindisease state: normal. (skin) | SRR040011(GSM532896) G529T. (cervix) | SRR330898(SRX091736) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191464(GSM715574) 112genomic small RNA (size selected RNA from . (breast) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | SRR038856(GSM458539) D11. (cell line) | SRR038857(GSM458540) D20. (cell line) | SRR191590(GSM715700) 48genomic small RNA (size selected RNA from t. (breast) | TAX577579(Rovira) total RNA. (breast) | SRR038858(GSM458541) MEL202. (cell line) | SRR038852(GSM458535) QF1160MB. (cell line) | SRR040037(GSM532922) G243T. (cervix) | DRR001483(DRX001037) Hela long cytoplasmic cell fraction, control. (hela) | DRR001485(DRX001039) Hela long total cell fraction, LNA(+). (hela) | SRR330914(SRX091752) tissue: normal skindisease state: normal. (skin) | SRR553574(SRX182780) source: Heart. (Heart) | TAX577738(Rovira) total RNA. (breast) | SRR029124(GSM416753) HeLa. (hela) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR330911(SRX091749) tissue: normal skindisease state: normal. (skin) | SRR191605(GSM715715) 76genomic small RNA (size selected RNA from t. (breast) | SRR363673(GSM830250) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR037937(GSM510475) 293cand2. (cell line) | SRR191434(GSM715544) 171genomic small RNA (size selected RNA from . (breast) | SRR189783 | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | GSM1105750AGO3(GSM1105750) small RNA sequencing data. (ago3 hela) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | DRR000558(DRX000316) THP-1 whole cell RNA, after 3 day treatment w. (cell line) | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) | SRR330885(SRX091723) tissue: skin psoriatic uninvolveddisease stat. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................................................................................................CAGCCTCCCCTCATCTGCT.......... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................CTTCACCCGCTTCTGATGT.......................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................CCTCCCTTTCCTCTCACC............................................................................................................................................................................. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................AACAGGGCTTCACCCGCGCT................................................................................ | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....CCTCCTCTCTTTCCCTGGGTG................................................................................................................................................................................................................................ | 21 | 1.00 | 0.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................CCATCGCTTGGACCCCTCCT.................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................TCTCTCTTCTGACCCCATCCTCCTAGG................................................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................GCCTCCCCTCATCTGG........... | 16 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - |
| .......................................................................................................................................................................................................................................TCATCTGGGATGTCA.... | 15 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 |