| (2) AGO2.ip | (1) BRAIN | (8) BREAST | (12) CELL-LINE | (1) FIBROBLAST | (2) HEART | (8) LIVER | (2) OTHER | (1) RRP40.ip | (10) SKIN | (1) UTERUS | (1) XRN.ip |
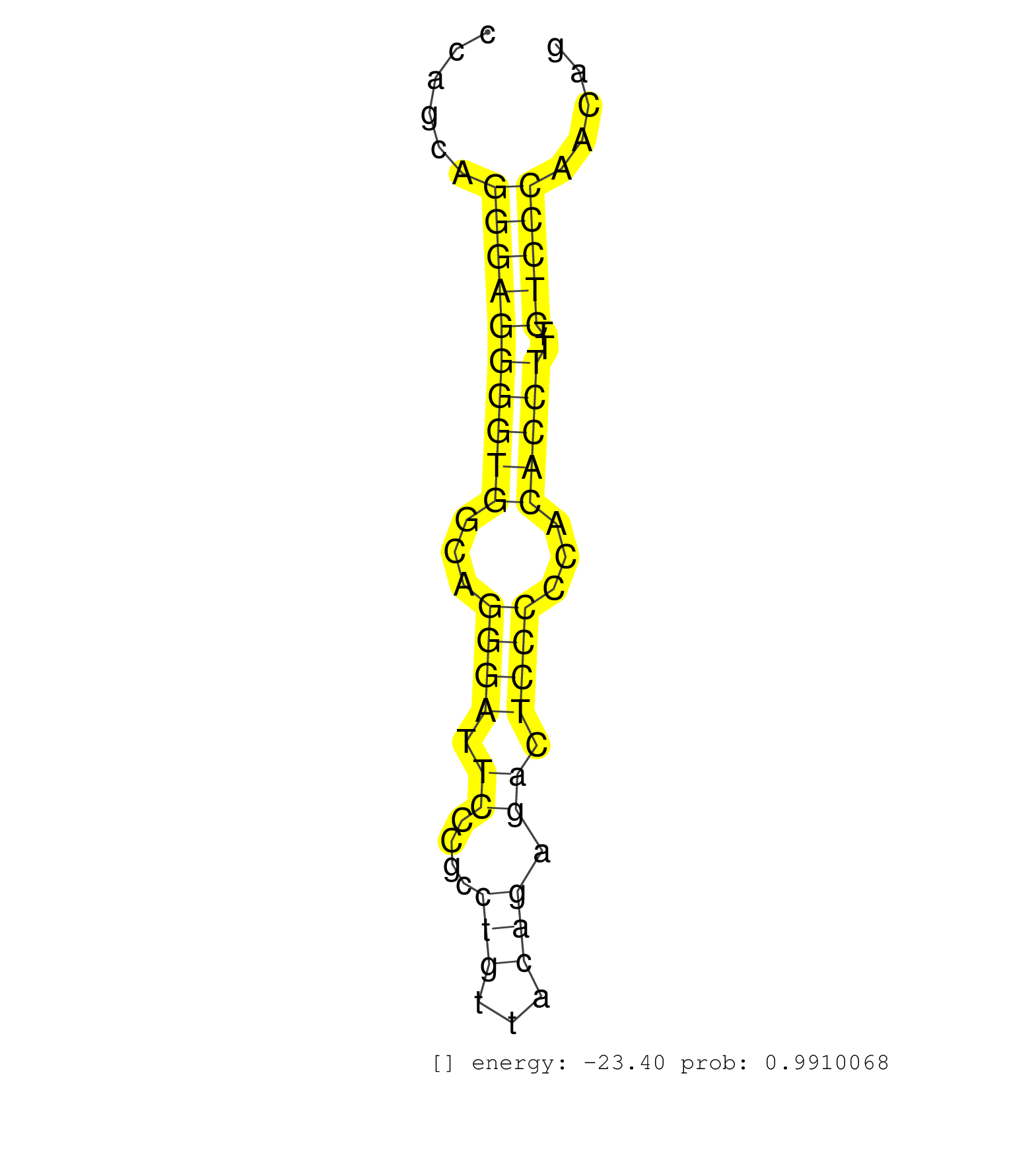
| CCGCAAGATGGGAGACAAGGTGGAGGCCCGGGCCATCGCCATTGCTGCGGGTGAATATAACGGGCAAGCAAGGGGTGGCCCCGCAGCTCCTGGAGAGGGTCCAGCAGGGAGGGGTGGCAGGGATTCCCGCCTGTTACAGAGACTCCCCCACACCTTTCTCCCAACAGGTGTTCCCGTTGTCCCTGGCACAGATGCCCCCATCACGTCCCTGCATGAG ..........................................................................................................((((((((((...((((.((....(((...))).)).))))...)))))..)))))....................................................... ....................................................................................................101...............................................................167................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR314796(SRX084354) Total RNA, fractionated (15-30nt). (cell line) | SRR189782 | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR207111(GSM721073) Whole cell RNA. (cell line) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR444053(SRX128901) Sample 13cDNABarcode: AF-PP-341: ACG CTC TTC . (skin) | SRR189783 | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | TAX577738(Rovira) total RNA. (breast) | TAX577746(Rovira) total RNA. (breast) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR038852(GSM458535) QF1160MB. (cell line) | SRR330900(SRX091738) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191574(GSM715684) 78genomic small RNA (size selected RNA from t. (breast) | SRR330892(SRX091730) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR060169(GSM565979) 5-8F_cytoplasm. (cell line) | SRR363676(GSM830253) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR037935(GSM510473) 293cand3. (cell line) | TAX577744(Rovira) total RNA. (breast) | SRR330904(SRX091742) tissue: normal skindisease state: normal. (skin) | DRR000557(DRX000315) THP-1 whole cell RNA, after 3 day treatment w. (cell line) | GSM956925Paz8D5(GSM956925) cell line: HEK293 cell linepassages: 15-20ip . (cell line) | SRR191622(GSM715732) 175genomic small RNA (size selected RNA from . (breast) | SRR038862(GSM458545) MM472. (cell line) | SRR039611(GSM531974) Human Normal Liver Tissue Sample 1. (liver) | SRR191491(GSM715601) 150genomic small RNA (size selected RNA from . (breast) | SRR191605(GSM715715) 76genomic small RNA (size selected RNA from t. (breast) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330872(SRX091710) tissue: skin psoriatic involveddisease state:. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................CTCCCCCACACCTTTCTCCCAAC.................................................... | 23 | 1 | 13.00 | 13.00 | 1.00 | - | 1.00 | 1.00 | 3.00 | - | - | 1.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - |
| ..............................................................................................................................................CTCCCCCACACCTTTCTCCCAACAG.................................................. | 25 | 1 | 8.00 | 8.00 | - | - | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................AGGGAGGGGTGGCAGGGATTATC......................................................................................... | 23 | 1 | 8.00 | 2.00 | - | 8.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................CAGGGAGGGGTGGCAGGGAT............................................................................................. | 20 | 1 | 4.00 | 4.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................CTCCCCCACACCTTTCTCCCAA..................................................... | 22 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ........................................................................................................................................................................................GGCACAGATGCCCCCATCACGT........... | 22 | 1 | 3.00 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................CTCCCCCACACCTTTCTCCCAACA................................................... | 24 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................CTCCCCCACACCTTTCTCCCA...................................................... | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................AGGGAGGGGTGGCAGGGATT............................................................................................ | 20 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................AGATGCCCCCATCACGTCCCT....... | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................AGGGAGGGGTGGCAGGGATTCGT......................................................................................... | 23 | 1 | 2.00 | 1.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................TCTCCCAACAGGTGTTCCGGT........................................ | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................................................CGTTGTCCCTGGCACAGATGCCCCCAT................ | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................CTCCCCCACACCTTTCTCCCAATA................................................... | 24 | 1 | 1.00 | 3.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................CCCATCACGTCCCTGCATGA. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................CTCCCCCACACCTTTCTCCCAACAGT................................................. | 26 | 1 | 1.00 | 8.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................CTCCCCCACACCTTTCTCCCAGAGT.................................................. | 25 | 1 | 1.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................AGGGAGGGGTGGCAGGGATTCA.......................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................CTCCCCCACACCTTTCTCCCAAT.................................................... | 23 | 1 | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................................................TTCCCGTTGTCCCTGGCACA........................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................TCCCCCACACCTTTCTCCCAAC.................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................AGGGAGGGGTGGCAGGGATTCAA......................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................AGGGAGGGGTGGCAGGGATTC........................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................AGGGAGGGGTGGCAGGGATTT........................................................................................... | 21 | 1 | 1.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................GGTGAATATAACGGGCAAGCAAGGGGTGGC.......................................................................................................................................... | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................AGGGAGGGGTGGCAGGGA.............................................................................................. | 18 | 3 | 1.00 | 1.00 | - | - | - | 0.33 | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - |
| ..............................................................................................................................................CTCCCCCACACCTTTCTCCCATA.................................................... | 23 | 1 | 1.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................AGGGAGGGGTGGCAGGGATTAAA......................................................................................... | 23 | 1 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................CAGGGAGGGGTGGCAGGGATTC........................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .CGCAAGATGGGAGACAAGG..................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ..........GGAGACAAGGTGGAGCAC............................................................................................................................................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................CAGGGAGGGGTGGCAGGGATAA........................................................................................... | 22 | 1 | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................GTTGTCCCTGGCACAGATGCCCCCATC............... | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............AGACAAGGTGGAGGCCCGGGC........................................................................................................................................................................................ | 21 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................CTCCCCCACACCTTTCTCCCATT.................................................... | 23 | 1 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................GCCCGGGCCATCGCC................................................................................................................................................................................. | 15 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..GCAAGATGGGAGACAAGGTG................................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................AGGGAGGGGTGGCAGGGATA............................................................................................ | 20 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................AGGGAGGGGTGGCAGGGAT............................................................................................. | 19 | 2 | 0.50 | 0.50 | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................CAGGGAGGGGTGGCAGGGA.............................................................................................. | 19 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - |
| .......ATGGGAGACAAGGTGGA................................................................................................................................................................................................. | 17 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................CAAGCAAGGGGTGGC.......................................................................................................................................... | 15 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 |
| .....................................................................................................CAGCAGGGAGGGGTGGC................................................................................................... | 17 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - |
| ........................................................................................................CAGGGAGGGGTGGCAGG................................................................................................ | 17 | 6 | 0.17 | 0.17 | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................AGGGAGGGGTGGCAGGG............................................................................................... | 17 | 7 | 0.14 | 0.14 | - | - | - | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CCGCAAGATGGGAGACAAGGTGGAGGCCCGGGCCATCGCCATTGCTGCGGGTGAATATAACGGGCAAGCAAGGGGTGGCCCCGCAGCTCCTGGAGAGGGTCCAGCAGGGAGGGGTGGCAGGGATTCCCGCCTGTTACAGAGACTCCCCCACACCTTTCTCCCAACAGGTGTTCCCGTTGTCCCTGGCACAGATGCCCCCATCACGTCCCTGCATGAG ..........................................................................................................((((((((((...((((.((....(((...))).)).))))...)))))..)))))....................................................... ....................................................................................................101...............................................................167................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR314796(SRX084354) Total RNA, fractionated (15-30nt). (cell line) | SRR189782 | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR207111(GSM721073) Whole cell RNA. (cell line) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR444053(SRX128901) Sample 13cDNABarcode: AF-PP-341: ACG CTC TTC . (skin) | SRR189783 | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR039619(GSM531982) HBV(+) HCC Tissue Sample 1. (liver) | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | TAX577738(Rovira) total RNA. (breast) | TAX577746(Rovira) total RNA. (breast) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR038852(GSM458535) QF1160MB. (cell line) | SRR330900(SRX091738) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191574(GSM715684) 78genomic small RNA (size selected RNA from t. (breast) | SRR330892(SRX091730) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR060169(GSM565979) 5-8F_cytoplasm. (cell line) | SRR363676(GSM830253) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR037935(GSM510473) 293cand3. (cell line) | TAX577744(Rovira) total RNA. (breast) | SRR330904(SRX091742) tissue: normal skindisease state: normal. (skin) | DRR000557(DRX000315) THP-1 whole cell RNA, after 3 day treatment w. (cell line) | GSM956925Paz8D5(GSM956925) cell line: HEK293 cell linepassages: 15-20ip . (cell line) | SRR191622(GSM715732) 175genomic small RNA (size selected RNA from . (breast) | SRR038862(GSM458545) MM472. (cell line) | SRR039611(GSM531974) Human Normal Liver Tissue Sample 1. (liver) | SRR191491(GSM715601) 150genomic small RNA (size selected RNA from . (breast) | SRR191605(GSM715715) 76genomic small RNA (size selected RNA from t. (breast) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330872(SRX091710) tissue: skin psoriatic involveddisease state:. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................CCCCGCAGCTCCTGGTGTC........................................................................................................................ | 19 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................CGCAGCTCCTGGAGA......................................................................................................................... | 15 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - |