| (1) AGO1.ip | (3) AGO2.ip | (4) B-CELL | (7) BRAIN | (10) BREAST | (21) CELL-LINE | (3) CERVIX | (5) HEART | (3) HELA | (2) LIVER | (3) OTHER | (1) RRP40.ip | (11) SKIN | (1) UTERUS | (1) XRN.ip |
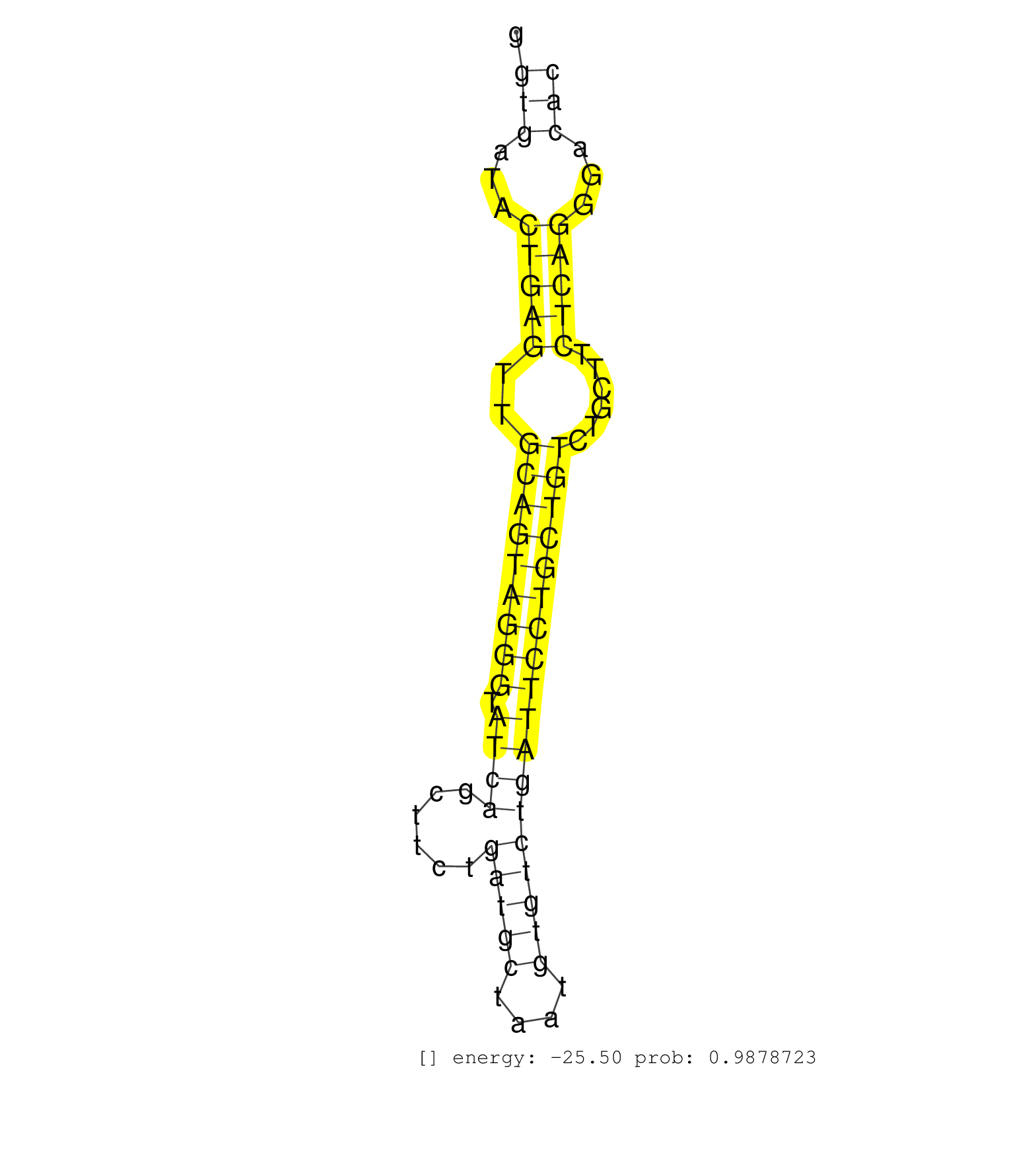
| AGTCTTACCCAGTATATATCTTTACATTTGCTTTTGCCATTGTATATACTTTCCGGTCCTATCTGAGCTCAGAGACTGGGCTTTCTTAATGATGCTCCGTTTCCCATTGTCATCATTAGAGCCTCTGTGGTGATACTGAGTTGCAGTAGGGTATCAGCTTCTGATGCTAATGTGTCTGATTCCTGCTGTCTGCTTCTCAGGGACACCATGTGGCGGATCTTTACTGGATCGCTGCTGGTAGAGGAGAAGT .................................................................................................................................(((...(((((..(((((((((.((((......(((((....))))))))))))))))))......)))))...)))............................................ ................................................................................................................................129..........................................................................206.......................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR037943(GSM510481) 293DcrTN. (cell line) | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR189782 | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | DRR001488(DRX001042) Hela short nuclear cell fraction, LNA(+). (hela) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR039191(GSM494810) PBMCs were isolated by ficoll gradient from t. (blood) | TAX577739(Rovira) total RNA. (breast) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | TAX577741(Rovira) total RNA. (breast) | SRR015446(SRR015446) smallRNAs high-throughput sequencing Total. (breast) | SRR330872(SRX091710) tissue: skin psoriatic involveddisease state:. (skin) | SRR040011(GSM532896) G529T. (cervix) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR330869(SRX091707) tissue: skin psoriatic involveddisease state:. (skin) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | DRR000561(DRX000319) Isolation of RNA following immunoprecipitatio. (ago2 cell line) | GSM450597(GSM450597) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450604(GSM450604) miRNA sequencing raw reads from post-mortem s. (brain) | SRR207119(GSM721081) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | GSM541797(GSM541797) differentiated human embryonic stem cells. (cell line) | SRR033720(GSM497065) EBV activated B cell line (EBV159). (B cell) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | SRR029131(GSM416760) MCF7. (cell line) | SRR039636(GSM518473) THP1_cyto_sRNAs. (cell line) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191624(GSM715734) 31genomic small RNA (size selected RNA from t. (breast) | TAX577744(Rovira) total RNA. (breast) | SRR037935(GSM510473) 293cand3. (cell line) | DRR001489(DRX001043) Hela short nuclear cell fraction, control. (hela) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR060986(GSM569190) Human memory B cell [09-001]. (cell line) | SRR033730(GSM497075) Lymphoblastic (ALL411). (B cell) | SRR033712(GSM497057) Burkitt Lymphoma (BL510). (B cell) | SRR191499(GSM715609) 6genomic small RNA (size selected RNA from to. (breast) | SRR189786 | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | SRR033719(GSM497064) 6hr Activated B cell line (ABC158). (B cell) | SRR040028(GSM532913) G026N. (cervix) | GSM532874(GSM532874) G699T. (cervix) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR330892(SRX091730) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM1105753INPUT(GSM1105753) small RNA sequencing data. (hela) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR038857(GSM458540) D20. (cell line) | SRR191550(GSM715660) 27genomic small RNA (size selected RNA from t. (breast) | SRR029125(GSM416754) U2OS. (cell line) | TAX577740(Rovira) total RNA. (breast) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | SRR191498(GSM715608) 3genomic small RNA (size selected RNA from to. (breast) | SRR444057(SRX128905) Sample 15cDNABarcode: AF-PP-334: ACG CTC TTC . (skin) | SRR314796(SRX084354) Total RNA, fractionated (15-30nt). (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................TACTGAGTTGCAGTAGGGTAT................................................................................................ | 21 | 1 | 16.00 | 16.00 | 1.00 | 2.00 | - | 1.00 | 1.00 | 1.00 | - | - | - | - | 2.00 | - | - | - | - | 1.00 | - | 2.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .....................................................................................................................................TACTGAGTTGCAGTAGGGTATC............................................................................................... | 22 | 1 | 14.00 | 14.00 | - | - | 4.00 | - | 1.00 | 1.00 | - | - | - | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................TGAGTTGCAGTAGGGTATCAGCT........................................................................................... | 23 | 1 | 10.00 | 10.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - |
| ..................................................................................................................................................................................ATTCCTGCTGTCTGCTTCTCAGA................................................. | 23 | 1 | 4.00 | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TACTGAGTTGCAGTAGGGTATCA.............................................................................................. | 23 | 1 | 4.00 | 4.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| ........................................................................................................................................TGAGTTGCAGTAGGGTATCA.............................................................................................. | 20 | 1 | 4.00 | 4.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................ATTCCTGCTGTCTGCTTCTCAGTT................................................ | 24 | 1 | 4.00 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TACTGAGTTGCAGTAGGGTAA................................................................................................ | 21 | 1 | 3.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................................GGATCGCTGCTGGTAGAGGAGA... | 22 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TACTGAGTTGCAGTAGGGTATCAG............................................................................................. | 24 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................ACTGGATCGCTGCTGGTAGAGGAG.... | 24 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TACTGAGTTGCAGTAGGGTATCT.............................................................................................. | 23 | 1 | 2.00 | 14.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................ACTGAGTTGCAGTAGGGTATCAGCTTCT........................................................................................ | 28 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................................GATCTTTACTGGATCGCTGCTGGTAG......... | 26 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................ACTGAGTTGCAGTAGGGTATCA.............................................................................................. | 22 | 1 | 2.00 | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................TGAGTTGCAGTAGGGTATCAGC............................................................................................ | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................TCTTTACTGGATCGCTGCTGGTAGA........ | 25 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TACTGAGTTGCAGTAGGGTATCAGA............................................................................................ | 25 | 1 | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TACTGAGTTGCAGTAGGGT.................................................................................................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................TGAGTTGCAGTAGGGTATCAGCTAGC........................................................................................ | 26 | 1 | 1.00 | 10.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................AGGGTATCAGCTTCTGATGCT.................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TACTGAGTTGCAGTAGGGTATCG.............................................................................................. | 23 | 1 | 1.00 | 14.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................................................................CGCTGCTGGTAGAGGAGA... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................ATTCCTGCTGTCTGCTTCTCAG.................................................. | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................................................................TTACTGGATCGCTGCTGGTAGAGGAG.... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| .....................................................CGGTCCTATCTGAGCTCAGAGA............................................................................................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................ATTCCTGCTGTCTGCTTCTCCGTT................................................ | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................TACTGAGTTGCAGTAGGGTATA............................................................................................... | 22 | 1 | 1.00 | 16.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| ................................................CTTTCCGGTCCTATCTGAGCTCAGAGAC.............................................................................................................................................................................. | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................GACACCATGTGGCGGATCTT............................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TACTGAGTTGCAGTAGGGGAT................................................................................................ | 21 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................ACTGAGTTGCAGTAGGGTAT................................................................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TTCCTGCTGTCTGCTTCTCAGTT................................................ | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | |
| ........................................................................................................................................TGAGTTGCAGTAGGGTATCAGCTT.......................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TACTGAGTTGCAGTAGGGTATCTT............................................................................................. | 24 | 1 | 1.00 | 14.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................................ACTGGATCGCTGCTGGTAGAGG...... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TACTGAGTTGCAGTAGGGTA................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................TGAGTTGCAGTAGGGTAGAA.............................................................................................. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................GTTGCAGTAGGGTATCGG............................................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................TGAGTTGCAGTAGGGTATCAGCA........................................................................................... | 23 | 1 | 1.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................TTAGAGCCTCTGTGGTGATACTGAGTTGCAGTAGGG................................................................................................... | 36 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TACTGAGTTGCAGTAGGGTATCAGCT........................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................TGATTCCTGCTGTCTGCTTCT..................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ......................................................................................................................................ACTGAGTTGCAGTAGGGTATCAGCT........................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................ACTGAGTTGCAGTAGGGTATCAGCTTTT........................................................................................ | 28 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................................................................................TGGATCGCTGCTGGTAGAGGAG.... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................CTGAGTTGCAGTAGGGTATCA.............................................................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................CTGGATCGCTGCTGGTAGAGG...... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TCCTGCTGTCTGCTTCTCAGTT................................................ | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................TACTGAGTTGCAGTAGGGTAAAG.............................................................................................. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................................................ATCGCTGCTGGTAGAGGAGAAGT | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................CTGGATCGCTGCTGGTAGAG....... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................TCCTGCTGTCTGCTTCTCATGA................................................ | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................................................................................CTTTACTGGATCGCTGCTGG............ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................ATTCCTGCTGTCTGCTTCT..................................................... | 19 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - |
| ....................................................................................................................................................................................................................................TCGCTGCTGGTAGA........ | 14 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........CAGTATATATCTTTAC................................................................................................................................................................................................................................. | 16 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - |
| AGTCTTACCCAGTATATATCTTTACATTTGCTTTTGCCATTGTATATACTTTCCGGTCCTATCTGAGCTCAGAGACTGGGCTTTCTTAATGATGCTCCGTTTCCCATTGTCATCATTAGAGCCTCTGTGGTGATACTGAGTTGCAGTAGGGTATCAGCTTCTGATGCTAATGTGTCTGATTCCTGCTGTCTGCTTCTCAGGGACACCATGTGGCGGATCTTTACTGGATCGCTGCTGGTAGAGGAGAAGT .................................................................................................................................(((...(((((..(((((((((.((((......(((((....))))))))))))))))))......)))))...)))............................................ ................................................................................................................................129..........................................................................206.......................................... | Size | Perfect hit | Total Norm | Perfect Norm | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR037943(GSM510481) 293DcrTN. (cell line) | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR189782 | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | DRR001488(DRX001042) Hela short nuclear cell fraction, LNA(+). (hela) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR039191(GSM494810) PBMCs were isolated by ficoll gradient from t. (blood) | TAX577739(Rovira) total RNA. (breast) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | TAX577741(Rovira) total RNA. (breast) | SRR015446(SRR015446) smallRNAs high-throughput sequencing Total. (breast) | SRR330872(SRX091710) tissue: skin psoriatic involveddisease state:. (skin) | SRR040011(GSM532896) G529T. (cervix) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR330869(SRX091707) tissue: skin psoriatic involveddisease state:. (skin) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | DRR000561(DRX000319) Isolation of RNA following immunoprecipitatio. (ago2 cell line) | GSM450597(GSM450597) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450604(GSM450604) miRNA sequencing raw reads from post-mortem s. (brain) | SRR207119(GSM721081) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | GSM541797(GSM541797) differentiated human embryonic stem cells. (cell line) | SRR033720(GSM497065) EBV activated B cell line (EBV159). (B cell) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | SRR029131(GSM416760) MCF7. (cell line) | SRR039636(GSM518473) THP1_cyto_sRNAs. (cell line) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191624(GSM715734) 31genomic small RNA (size selected RNA from t. (breast) | TAX577744(Rovira) total RNA. (breast) | SRR037935(GSM510473) 293cand3. (cell line) | DRR001489(DRX001043) Hela short nuclear cell fraction, control. (hela) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR060986(GSM569190) Human memory B cell [09-001]. (cell line) | SRR033730(GSM497075) Lymphoblastic (ALL411). (B cell) | SRR033712(GSM497057) Burkitt Lymphoma (BL510). (B cell) | SRR191499(GSM715609) 6genomic small RNA (size selected RNA from to. (breast) | SRR189786 | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | SRR033719(GSM497064) 6hr Activated B cell line (ABC158). (B cell) | SRR040028(GSM532913) G026N. (cervix) | GSM532874(GSM532874) G699T. (cervix) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR330892(SRX091730) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM1105753INPUT(GSM1105753) small RNA sequencing data. (hela) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR038857(GSM458540) D20. (cell line) | SRR191550(GSM715660) 27genomic small RNA (size selected RNA from t. (breast) | SRR029125(GSM416754) U2OS. (cell line) | TAX577740(Rovira) total RNA. (breast) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | SRR191498(GSM715608) 3genomic small RNA (size selected RNA from to. (breast) | SRR444057(SRX128905) Sample 15cDNABarcode: AF-PP-334: ACG CTC TTC . (skin) | SRR314796(SRX084354) Total RNA, fractionated (15-30nt). (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................................ATCAGCTTCTGATGCTTTG............................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................GATGCTCCGTTTCC.................................................................................................................................................. | 14 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 |