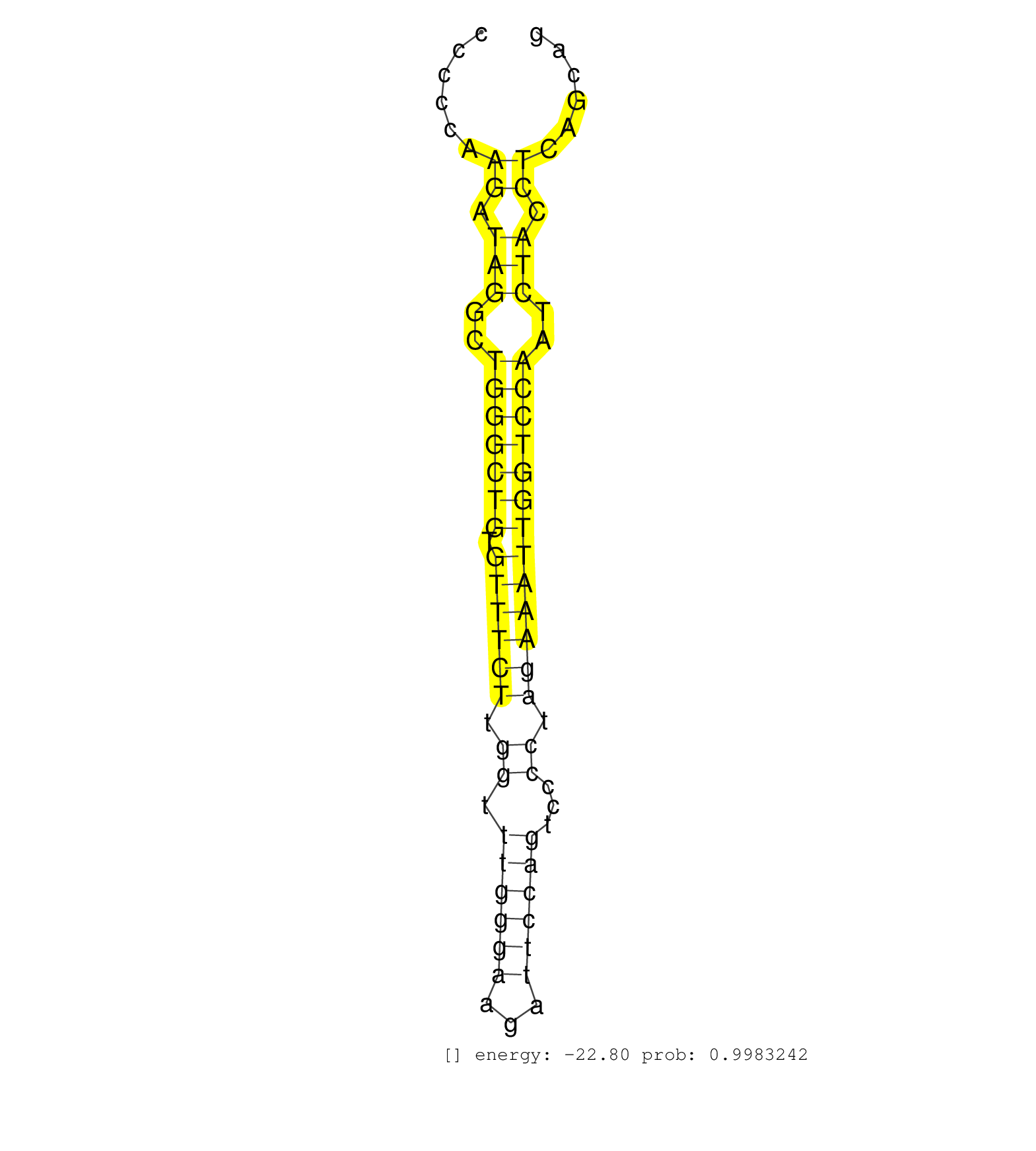
| (3) AGO1.ip | (1) AGO1.ip OTHER.mut | (4) AGO2.ip | (1) AGO3.ip | (16) B-CELL | (12) BRAIN | (14) BREAST | (55) CELL-LINE | (6) CERVIX | (2) FIBROBLAST | (8) HEART | (5) HELA | (1) KIDNEY | (8) LIVER | (2) OTHER | (1) RRP40.ip | (45) SKIN | (1) TESTES | (4) UTERUS | (1) XRN.ip |
| CAGGATGCCAATGCCTCTTCCCTCTTAGACATCTATAGCTTCTGGCTCAAGTAAGCCTTTCCTGTTCCATTTTGGCTATTTTCTCCCCCAAGATAGGCTGGGCTGTGTTTCTTGGTTTGGGAAGATTCCAGTCCCCTAGAAATTGGTCCAATCTACCTCAGCAGGTCTGCCAAGGTCCCAGAGCGAAAGTTACAGGCAAATGGACCAGTGGCTA ..........................................................................................((.(((..(((((((.((((((.((.((((((...))))))...)).)))))))))))))..))).))........................................................ ....................................................................................85.............................................................................164................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR189782 | SRR387909(GSM843861) specific-host: Homo sapienshost cell line: Su. (cell line) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039193(GSM494812) HL60 cell line is derived from acute promyelo. (cell line) | SRR033711(GSM497056) GCB DLBCL (GCB110). (B cell) | SRR189786 | DRR000557(DRX000315) THP-1 whole cell RNA, after 3 day treatment w. (cell line) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR037942(GSM510480) 293DroshaTN_cand5. (cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR037938(GSM510476) 293Red. (cell line) | SRR033712(GSM497057) Burkitt Lymphoma (BL510). (B cell) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR038858(GSM458541) MEL202. (cell line) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR038863(GSM458546) MM603. (cell line) | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | SRR390723(GSM850202) total small RNA. (cell line) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | GSM1105748AGO1(GSM1105748) small RNA sequencing data. (ago1 hela) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | SRR060983(GSM569187) Human pre-germinal center B cell [09-001]. (cell line) | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | DRR001489(DRX001043) Hela short nuclear cell fraction, control. (hela) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039624(GSM531987) HBV(-) HCV(-) Adjacent Tissue Sample. (liver) | SRR553574(SRX182780) source: Heart. (Heart) | SRR189787 | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | GSM1105750AGO3(GSM1105750) small RNA sequencing data. (ago3 hela) | SRR039191(GSM494810) PBMCs were isolated by ficoll gradient from t. (blood) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | SRR033723(GSM497068) L1236 cell line (L1236). (B cell) | SRR029128(GSM416757) H520. (cell line) | SRR060169(GSM565979) 5-8F_cytoplasm. (cell line) | SRR207114(GSM721076) IP against AGO 1 & 2, RRP40 knockdown. (ago1/2 RRP40 cell line) | SRR189784 | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | SRR326282(GSM769512) Dicer mRNA was knocked down using siDicer, to. (cell line) | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577739(Rovira) total RNA. (breast) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR038861(GSM458544) MM466. (cell line) | SRR033716(GSM497061) Mentle Cell Lymphoma (MCL114). (B cell) | SRR037941(GSM510479) 293DroshaTN. (cell line) | SRR326281(GSM769511) Dicer mRNA was knocked down using siDicer, cy. (cell line) | SRR343332(GSM796035) KSHV (HHV8), EBV (HHV-4). (cell line) | SRR033715(GSM497060) Mantle Cell Lymphoma (Mino122). (B cell) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR343333(GSM796036) KSHV (HHV8). (cell line) | SRR330903(SRX091741) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | DRR000556(DRX000314) THP-1 whole cell RNA, after 3 day treatment w. (cell line) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | SRR039637(GSM518474) THP1_total_sRNAs. (cell line) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR330888(SRX091726) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | SRR330913(SRX091751) tissue: normal skindisease state: normal. (skin) | SRR330914(SRX091752) tissue: normal skindisease state: normal. (skin) | GSM450603(GSM450603) miRNA sequencing raw reads from post-mortem s. (brain) | SRR060985(GSM569189) Human naive B cell [09-001]. (cell line) | TAX577746(Rovira) total RNA. (breast) | SRR207111(GSM721073) Whole cell RNA. (cell line) | DRR001486(DRX001040) Hela long cytoplasmic cell fraction, LNA(+). (hela) | SRR038862(GSM458545) MM472. (cell line) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | DRR000558(DRX000316) THP-1 whole cell RNA, after 3 day treatment w. (cell line) | SRR037937(GSM510475) 293cand2. (cell line) | TAX577744(Rovira) total RNA. (breast) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450598(GSM450598) miRNA sequencing raw reads from post-mortem s. (brain) | DRR000559(DRX000317) THP-1 whole cell RNA, no treatment. (cell line) | SRR553575(SRX182781) source: Kidney. (Kidney) | DRR000560(DRX000318) Isolation of RNA following immunoprecipitatio. (ago1 cell line) | GSM450607(GSM450607) miRNA sequencing raw reads from post-mortem s. (brain) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR060984(GSM569188) Human plasma cell [09-001]. (cell line) | SRR553576(SRX182782) source: Testis. (testes) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | SRR033717(GSM497062) Mentle Cell Lymphoma (MCL112). (B cell) | SRR015359(GSM380324) Germinal Center B cell (GC136). (B cell) | SRR330872(SRX091710) tissue: skin psoriatic involveddisease state:. (skin) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | SRR039635(GSM518472) THP1_nuc_sRNAs. (cell line) | SRR038852(GSM458535) QF1160MB. (cell line) | DRR000561(DRX000319) Isolation of RNA following immunoprecipitatio. (ago2 cell line) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | TAX577453(Rovira) total RNA. (breast) | SRR037939(GSM510477) 293cand5_rep1. (cell line) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | GSM450599(GSM450599) miRNA sequencing raw reads from post-mortem s. (brain) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | GSM359177(GSM359177) hela_nucl_a. (hela) | GSM956925AGO2Paz8(GSM956925) cell line: HEK293 cell linepassages: 15-20ip . (ago2 cell line) | SRR033730(GSM497075) Lymphoblastic (ALL411). (B cell) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR033728(GSM497073) MALT (MALT413). (B cell) | DRR000555(DRX000313) THP-1 whole cell RNA, no treatment. (cell line) | SRR330875(SRX091713) tissue: skin psoriatic involveddisease state:. (skin) | TAX577740(Rovira) total RNA. (breast) | SRR191585(GSM715695) 196genomic small RNA (size selected RNA from . (breast) | GSM532876(GSM532876) G547T. (cervix) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) | TAX577589(Rovira) total RNA. (breast) | SRR330859(SRX091697) tissue: skin psoriatic involveddisease state:. (skin) | SRR343335 | TAX577743(Rovira) total RNA. (breast) | SRR330916(SRX091754) tissue: normal skindisease state: normal. (skin) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577745(Rovira) total RNA. (breast) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR330883(SRX091721) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | SRR330865(SRX091703) tissue: skin psoriatic involveddisease state:. (skin) | SRR330894(SRX091732) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330899(SRX091737) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330880(SRX091718) tissue: skin psoriatic involveddisease state:. (skin) | SRR330891(SRX091729) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR363673(GSM830250) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | SRR029132(GSM416761) MB-MDA231. (cell line) | TAX577741(Rovira) total RNA. (breast) | SRR330874(SRX091712) tissue: skin psoriatic involveddisease state:. (skin) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR033726(GSM497071) Mututated CLL (CLLM633). (B cell) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR029129(GSM416758) SW480. (cell line) | SRR330860(SRX091698) tissue: skin psoriatic involveddisease state:. (skin) | SRR444058(SRX128906) Sample 16cDNABarcode: AF-PP-335: ACG CTC TTC . (skin) | SRR330923(SRX091761) tissue: normal skindisease state: normal. (skin) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | SRR191601(GSM715711) 58genomic small RNA (size selected RNA from t. (breast) | SRR444060(SRX128908) Sample 18cDNABarcode: AF-PP-340: ACG CTC TTC . (skin) | SRR039636(GSM518473) THP1_cyto_sRNAs. (cell line) | SRR040015(GSM532900) G623T. (cervix) | SRR040028(GSM532913) G026N. (cervix) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR040031(GSM532916) G013T. (cervix) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) | SRR444049(SRX128897) Sample 9cDNABarcode: AF-PP-334: ACG CTC TTC C. (skin) | SRR040010(GSM532895) G529N. (cervix) | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR330886(SRX091724) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039611(GSM531974) Human Normal Liver Tissue Sample 1. (liver) | SRR363674(GSM830251) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR040018(GSM532903) G701N. (cervix) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | TAX577580(Rovira) total RNA. (breast) | SRR330900(SRX091738) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330885(SRX091723) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191471(GSM715581) 119genomic small RNA (size selected RNA from . (breast) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................AAGATAGGCTGGGCTGTGTTTCT...................................................................................................... | 23 | 1 | 78.00 | 78.00 | 13.00 | - | - | 3.00 | 1.00 | 4.00 | 2.00 | 3.00 | 1.00 | 3.00 | 3.00 | - | 1.00 | 1.00 | 1.00 | - | 4.00 | - | - | 2.00 | 2.00 | 4.00 | - | - | 1.00 | - | 2.00 | 1.00 | - | - | 1.00 | 1.00 | - | - | 1.00 | - | 1.00 | - | - | 1.00 | 2.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | 1.00 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | 1.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | 1.00 | - | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................AAGATAGGCTGGGCTGTGTTTC....................................................................................................... | 22 | 1 | 55.00 | 55.00 | 12.00 | - | 8.00 | 2.00 | 2.00 | - | 2.00 | 1.00 | - | - | 2.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | 1.00 | - | 1.00 | - | - | - | 1.00 | - | - | 1.00 | 1.00 | - | - | 1.00 | - | 1.00 | 1.00 | - | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................AAGATAGGCTGGGCTGTGTTT........................................................................................................ | 21 | 1 | 45.00 | 45.00 | 4.00 | - | - | 5.00 | - | - | 1.00 | 1.00 | - | 1.00 | - | 3.00 | - | - | 1.00 | - | - | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | 1.00 | 2.00 | - | - | - | 1.00 | - | 1.00 | - | 2.00 | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | 1.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................AAGATAGGCTGGGCTGTGTT......................................................................................................... | 20 | 1 | 16.00 | 16.00 | - | - | 5.00 | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................AAATTGGTCCAATCTACCTCAG..................................................... | 22 | 1 | 11.00 | 11.00 | - | 8.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................ATAGGCTGGGCTGTGTTTCTTGG................................................................................................... | 23 | 1 | 6.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | 4.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................CTTAGACATCTATAGCTTCTGGC........................................................................................................................................................................ | 23 | 1 | 5.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................AAGATAGGCTGGGCTGTGTTTAA...................................................................................................... | 23 | 1 | 4.00 | 45.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................CAAGATAGGCTGGGCTGTGTTT........................................................................................................ | 22 | 1 | 4.00 | 4.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................AAGATAGGCTGGGCTGTGTTTCTT..................................................................................................... | 24 | 1 | 4.00 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................AGATAGGCTGGGCTGTGTTTCT...................................................................................................... | 22 | 1 | 3.00 | 3.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................AAGATAGGCTGGGCTGTGTTA........................................................................................................ | 21 | 1 | 3.00 | 16.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................AAGATAGGCTGGGCTGTGT.......................................................................................................... | 19 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................AAGATAGGCTGGGCTGTGTTTA....................................................................................................... | 22 | 1 | 3.00 | 45.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................AAGATAGGCTGGGCTGTGTTTT....................................................................................................... | 22 | 1 | 3.00 | 45.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................AGGTCCCAGAGCGAAAGTT....................... | 19 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................AGATAGGCTGGGCTGTGTTT........................................................................................................ | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................CAAGATAGGCTGGGCTGTGTTTC....................................................................................................... | 23 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................ATAGGCTGGGCTGTGTTTCTTG.................................................................................................... | 22 | 1 | 2.00 | 2.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................AAGATAGGCTGGGCTGTGTTTTATG.................................................................................................... | 25 | 1 | 2.00 | 45.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................AAGATAGGCTGGGCTGTGTTTTT...................................................................................................... | 23 | 1 | 2.00 | 45.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................AAGATAGGCTGGGCTGTGTTAA....................................................................................................... | 22 | 1 | 2.00 | 16.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................AAGATAGGCTGGGCTGGGT.......................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................AAATTGGTCCAATCTACCACAC..................................................... | 22 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................ATAGGCTGGGCTGTGTTTCTTGGT.................................................................................................. | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................ACATCTATAGCTTCTGGCTCAACAG................................................................................................................................................................. | 25 | 1.00 | 0.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................AAGATAGGCTGGGCTGGGTT......................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................................CTGCCAAGGTCCCAGAGCGAAAGTTACAGGCAAATG............ | 36 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................AAGATAGGCTGGGCTGTGGTT........................................................................................................ | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................AGACATCTATAGCTTCTGGCTCAAGTAA................................................................................................................................................................ | 28 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................TACAGGCAAATGGACCAGTGGCT. | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................AAATTGGTCCAATCTACCTCAA..................................................... | 22 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................AAGATAGGCTGGGCTGTGTTTCTAA.................................................................................................... | 25 | 1 | 1.00 | 78.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................CCAGAGCGAAAGTTACAGGCA................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................AAGATAGGCTGGGCTGTGTAA........................................................................................................ | 21 | 1 | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................TCTGCCAAGGTCCCAGAGCGA............................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................................GCAAATGGACCAGTGGCT. | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................GATAGGCTGGGCTGTGTTTCTT..................................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................AAGATAGGCTGGGCTGTGTTTCC...................................................................................................... | 23 | 1 | 1.00 | 55.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................ATAGGCTGGGCTGTGTTT........................................................................................................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................AAATTGGTCCAATCTACCTCAGA.................................................... | 23 | 1 | 1.00 | 11.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................TAGAAATTGGTCCAATCTACCT........................................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................GATAGGCTGGGCTGTGTTTCT...................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................AAGATAGGCTGGGCTGTGTTTAATG.................................................................................................... | 25 | 1 | 1.00 | 45.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................AGACATCTATAGCTTCTG.......................................................................................................................................................................... | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................AGATAGGCTGGGCTGTGTTTA....................................................................................................... | 21 | 1 | 1.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................AAGATAGGCTGGGCTGTGTTTAT...................................................................................................... | 23 | 1 | 1.00 | 45.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................................CAAATGGACCAGTGGCTA | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................ATAGGCTGGGCTGTGTTTC....................................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................AAATTGGTCCAATCTACCTCA...................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................ATAGGCTGGGCTGTGTTTAG...................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................CCAAGATAGGCTGGGCTGTGTTTC....................................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................CTATAGCTTCTGGCTCAAGTCTGC.............................................................................................................................................................. | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................GATAGGCTGGGCTGTGTTTCTTG.................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................CAGAGCGAAAGTTACAGGC................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................AGATAGGCTGGGCTGTGTTTC....................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........ATGCCTCTTCCCTCTTAGACATCTACAGC............................................................................................................................................................................... | 29 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................ATAGGCTGGGCTGTGTTTCCTGG................................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................CCAAGGTCCCAGAGCGAAAGTTACAG................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................TTAGACATCTATAGCTTCTGGCTCAA.................................................................................................................................................................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................CAAGATAGGCTGGGCTGTGTCTC....................................................................................................... | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................................................................AGTTACAGGCAAATGGACCAGTGGC.. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................AAATTGGTCCAATCTACCTAAG..................................................... | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................AAGATAGGCTGGGCTGT............................................................................................................ | 17 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................AAAGTTACAGGCAAATGGACCAGTGGCTA | 29 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................ATAGGCTGGGCTGTGTTTCTTGGA.................................................................................................. | 24 | 1 | 1.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................AAAGTTACAGGCAAATGGACCAGTG.... | 25 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................AAAGTTACAGGCAAATGGACC........ | 21 | 2 | 0.50 | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................CAATCTACCTCAGCAG.................................................. | 16 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................GGCAAATGGACCAGTGGC.. | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................................................AGCGAAAGTTACAGGCAAATGGACCA....... | 26 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................AGTTACAGGCAAATGGACC........ | 19 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................AAAGTTACAGGCAAATGGACCA....... | 22 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................AAAGTTACAGGCAAATGGACCAG...... | 23 | 2 | 0.50 | 0.50 | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................CCAGAGCGAAAGT........................ | 13 | 10 | 0.10 | 0.10 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.10 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CAGGATGCCAATGCCTCTTCCCTCTTAGACATCTATAGCTTCTGGCTCAAGTAAGCCTTTCCTGTTCCATTTTGGCTATTTTCTCCCCCAAGATAGGCTGGGCTGTGTTTCTTGGTTTGGGAAGATTCCAGTCCCCTAGAAATTGGTCCAATCTACCTCAGCAGGTCTGCCAAGGTCCCAGAGCGAAAGTTACAGGCAAATGGACCAGTGGCTA ..........................................................................................((.(((..(((((((.((((((.((.((((((...))))))...)).)))))))))))))..))).))........................................................ ....................................................................................85.............................................................................164................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR189782 | SRR387909(GSM843861) specific-host: Homo sapienshost cell line: Su. (cell line) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039193(GSM494812) HL60 cell line is derived from acute promyelo. (cell line) | SRR033711(GSM497056) GCB DLBCL (GCB110). (B cell) | SRR189786 | DRR000557(DRX000315) THP-1 whole cell RNA, after 3 day treatment w. (cell line) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR094129(GSM651905) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR037942(GSM510480) 293DroshaTN_cand5. (cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR037938(GSM510476) 293Red. (cell line) | SRR033712(GSM497057) Burkitt Lymphoma (BL510). (B cell) | SRR037940(GSM510478) 293cand5_rep2. (cell line) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR038858(GSM458541) MEL202. (cell line) | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR038863(GSM458546) MM603. (cell line) | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | SRR390723(GSM850202) total small RNA. (cell line) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | GSM1105748AGO1(GSM1105748) small RNA sequencing data. (ago1 hela) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | SRR060983(GSM569187) Human pre-germinal center B cell [09-001]. (cell line) | SRR033721(GSM497066) Ly3 cell line (Ly3). (B cell) | DRR001489(DRX001043) Hela short nuclear cell fraction, control. (hela) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039624(GSM531987) HBV(-) HCV(-) Adjacent Tissue Sample. (liver) | SRR553574(SRX182780) source: Heart. (Heart) | SRR189787 | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | GSM1105750AGO3(GSM1105750) small RNA sequencing data. (ago3 hela) | SRR039191(GSM494810) PBMCs were isolated by ficoll gradient from t. (blood) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | SRR033723(GSM497068) L1236 cell line (L1236). (B cell) | SRR029128(GSM416757) H520. (cell line) | SRR060169(GSM565979) 5-8F_cytoplasm. (cell line) | SRR207114(GSM721076) IP against AGO 1 & 2, RRP40 knockdown. (ago1/2 RRP40 cell line) | SRR189784 | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | SRR326282(GSM769512) Dicer mRNA was knocked down using siDicer, to. (cell line) | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577739(Rovira) total RNA. (breast) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR038861(GSM458544) MM466. (cell line) | SRR033716(GSM497061) Mentle Cell Lymphoma (MCL114). (B cell) | SRR037941(GSM510479) 293DroshaTN. (cell line) | SRR326281(GSM769511) Dicer mRNA was knocked down using siDicer, cy. (cell line) | SRR343332(GSM796035) KSHV (HHV8), EBV (HHV-4). (cell line) | SRR033715(GSM497060) Mantle Cell Lymphoma (Mino122). (B cell) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR343333(GSM796036) KSHV (HHV8). (cell line) | SRR330903(SRX091741) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | DRR000556(DRX000314) THP-1 whole cell RNA, after 3 day treatment w. (cell line) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | SRR039637(GSM518474) THP1_total_sRNAs. (cell line) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR330888(SRX091726) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | SRR330913(SRX091751) tissue: normal skindisease state: normal. (skin) | SRR330914(SRX091752) tissue: normal skindisease state: normal. (skin) | GSM450603(GSM450603) miRNA sequencing raw reads from post-mortem s. (brain) | SRR060985(GSM569189) Human naive B cell [09-001]. (cell line) | TAX577746(Rovira) total RNA. (breast) | SRR207111(GSM721073) Whole cell RNA. (cell line) | DRR001486(DRX001040) Hela long cytoplasmic cell fraction, LNA(+). (hela) | SRR038862(GSM458545) MM472. (cell line) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | DRR000558(DRX000316) THP-1 whole cell RNA, after 3 day treatment w. (cell line) | SRR037937(GSM510475) 293cand2. (cell line) | TAX577744(Rovira) total RNA. (breast) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450598(GSM450598) miRNA sequencing raw reads from post-mortem s. (brain) | DRR000559(DRX000317) THP-1 whole cell RNA, no treatment. (cell line) | SRR553575(SRX182781) source: Kidney. (Kidney) | DRR000560(DRX000318) Isolation of RNA following immunoprecipitatio. (ago1 cell line) | GSM450607(GSM450607) miRNA sequencing raw reads from post-mortem s. (brain) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR060984(GSM569188) Human plasma cell [09-001]. (cell line) | SRR553576(SRX182782) source: Testis. (testes) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | SRR033717(GSM497062) Mentle Cell Lymphoma (MCL112). (B cell) | SRR015359(GSM380324) Germinal Center B cell (GC136). (B cell) | SRR330872(SRX091710) tissue: skin psoriatic involveddisease state:. (skin) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | SRR039635(GSM518472) THP1_nuc_sRNAs. (cell line) | SRR038852(GSM458535) QF1160MB. (cell line) | DRR000561(DRX000319) Isolation of RNA following immunoprecipitatio. (ago2 cell line) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | TAX577453(Rovira) total RNA. (breast) | SRR037939(GSM510477) 293cand5_rep1. (cell line) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | GSM450599(GSM450599) miRNA sequencing raw reads from post-mortem s. (brain) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | GSM359177(GSM359177) hela_nucl_a. (hela) | GSM956925AGO2Paz8(GSM956925) cell line: HEK293 cell linepassages: 15-20ip . (ago2 cell line) | SRR033730(GSM497075) Lymphoblastic (ALL411). (B cell) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR033728(GSM497073) MALT (MALT413). (B cell) | DRR000555(DRX000313) THP-1 whole cell RNA, no treatment. (cell line) | SRR330875(SRX091713) tissue: skin psoriatic involveddisease state:. (skin) | TAX577740(Rovira) total RNA. (breast) | SRR191585(GSM715695) 196genomic small RNA (size selected RNA from . (breast) | GSM532876(GSM532876) G547T. (cervix) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) | TAX577589(Rovira) total RNA. (breast) | SRR330859(SRX091697) tissue: skin psoriatic involveddisease state:. (skin) | SRR343335 | TAX577743(Rovira) total RNA. (breast) | SRR330916(SRX091754) tissue: normal skindisease state: normal. (skin) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577745(Rovira) total RNA. (breast) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR330883(SRX091721) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | SRR330865(SRX091703) tissue: skin psoriatic involveddisease state:. (skin) | SRR330894(SRX091732) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330899(SRX091737) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330880(SRX091718) tissue: skin psoriatic involveddisease state:. (skin) | SRR330891(SRX091729) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR363673(GSM830250) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | SRR029132(GSM416761) MB-MDA231. (cell line) | TAX577741(Rovira) total RNA. (breast) | SRR330874(SRX091712) tissue: skin psoriatic involveddisease state:. (skin) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR033726(GSM497071) Mututated CLL (CLLM633). (B cell) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR029129(GSM416758) SW480. (cell line) | SRR330860(SRX091698) tissue: skin psoriatic involveddisease state:. (skin) | SRR444058(SRX128906) Sample 16cDNABarcode: AF-PP-335: ACG CTC TTC . (skin) | SRR330923(SRX091761) tissue: normal skindisease state: normal. (skin) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | SRR191601(GSM715711) 58genomic small RNA (size selected RNA from t. (breast) | SRR444060(SRX128908) Sample 18cDNABarcode: AF-PP-340: ACG CTC TTC . (skin) | SRR039636(GSM518473) THP1_cyto_sRNAs. (cell line) | SRR040015(GSM532900) G623T. (cervix) | SRR040028(GSM532913) G026N. (cervix) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | SRR040031(GSM532916) G013T. (cervix) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) | SRR444049(SRX128897) Sample 9cDNABarcode: AF-PP-334: ACG CTC TTC C. (skin) | SRR040010(GSM532895) G529N. (cervix) | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR330886(SRX091724) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039611(GSM531974) Human Normal Liver Tissue Sample 1. (liver) | SRR363674(GSM830251) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR040018(GSM532903) G701N. (cervix) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | TAX577580(Rovira) total RNA. (breast) | SRR330900(SRX091738) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330885(SRX091723) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191471(GSM715581) 119genomic small RNA (size selected RNA from . (breast) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .AGGATGCCAATGCCTAAG................................................................................................................................................................................................... | 18 | 14.17 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | 0.33 | 0.17 | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.33 | 0.33 | 0.33 | - | - | - | 0.17 | 0.17 | 1.00 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.83 | 0.33 | - | 0.50 | 0.50 | - | 0.17 | 0.33 | - | 0.17 | - | 0.50 | 0.17 | 0.50 | 0.17 | 0.33 | 0.17 | 0.33 | - | - | 0.17 | 0.33 | 0.17 | 0.17 | 0.33 | - | - | - | 0.17 | - | 0.17 | 0.17 | - | 0.17 | 0.17 | 0.17 | 0.17 | 0.17 | 0.17 | 0.17 | - | 0.17 | 0.17 | - | - | 0.17 | 0.17 | - | - | 0.17 | 0.17 | - | 0.17 | - | - | 0.17 | 0.17 | - | 0.17 | 0.17 | - | |
| .AGGATGCCAATGCCT...................................................................................................................................................................................................... | 15 | 6 | 5.50 | 5.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | 0.17 | - | - | - | 0.17 | - | - | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | 0.17 | 0.33 | - | 0.33 | - | 0.33 | - | 0.17 | - | 0.17 | - | 0.33 | 0.33 | - | - | 0.17 | 0.17 | - | - | 0.33 | - | - | 0.17 | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | 0.17 | 0.17 | - | - | 0.17 | 0.17 | - | - | 0.17 | - | 0.17 | 0.17 | - | - | 0.17 | - | - | - |
| .................................................................................................................................................GTCCAATCTACCTCAAGTT.................................................. | 19 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .AGGATGCCAATGCCTAAGC.................................................................................................................................................................................................. | 19 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | 0.17 | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | 0.17 | 0.17 | 0.33 | - | 0.17 | 0.33 | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .AGGATGCCAATGCCTAA.................................................................................................................................................................................................... | 17 | 1.17 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | 0.17 | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | |
| ..............................................................TGTTCCATTTTGGCTAAT...................................................................................................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................GGTCCAATCTACCTCAGCT................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................CAATCTACCTCAGCAAAA................................................ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................CAATCTACCTCAGCATA................................................. | 17 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................GCAGGTCTGCCAAGG....................................... | 15 | 3 | 0.67 | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................TATAGCTTCTGGCTCA..................................................................................................................................................................... | 16 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .AGGATGCCAATGCCTAATA.................................................................................................................................................................................................. | 19 | 0.17 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |