| (1) AGO1.ip | (2) AGO2.ip | (6) B-CELL | (15) BRAIN | (6) BREAST | (18) CELL-LINE | (1) CERVIX | (1) HEART | (4) LIVER | (2) OTHER | (6) SKIN | (2) UTERUS | (1) XRN.ip |
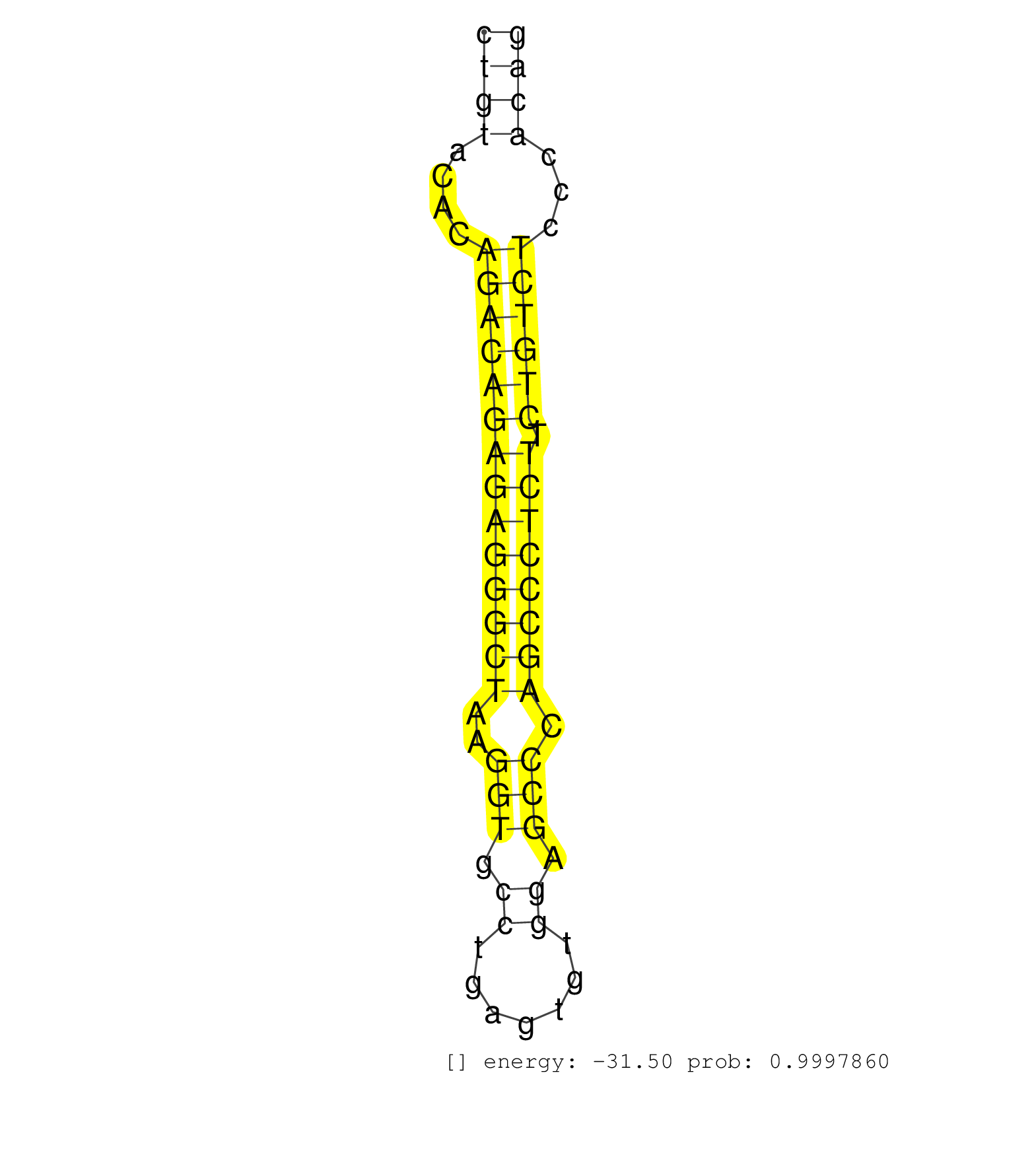
| TGCCCTTGTGGAGAAAGCAGTTTTCACTGCAACCTCTGGGGAGGGGAGCTCAACCCTGGGTCCCACTTTATCAAGAGTTTGGCTGCATGAGAATCACCCTGAAGCCCAGGCTTTCAACTACAGGGCTAGCCAGCCTGTACACAGACAGAGAGGGCTAAGGTGCCTGAGTGTGGAGCCCAGCCCTCTTCTGTCTCCCACAGTTTCAGATCCGAGCTCTGGAGTCCCAGAAGCGGCAGCAGGAGATGGTCCT ......................................................................................................................................((((....((((((((((((((..(((.((.......)).))).)))))))).))))))...)))).................................................. ......................................................................................................................................135..............................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | GSM339996(GSM339996) hues6Neuron. (cell line) | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450598(GSM450598) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450607(GSM450607) miRNA sequencing raw reads from post-mortem s. (brain) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | GSM450597(GSM450597) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039193(GSM494812) HL60 cell line is derived from acute promyelo. (cell line) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | GSM450599(GSM450599) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR060983(GSM569187) Human pre-germinal center B cell [09-001]. (cell line) | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR060985(GSM569189) Human naive B cell [09-001]. (cell line) | GSM450603(GSM450603) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039636(GSM518473) THP1_cyto_sRNAs. (cell line) | SRR191406(GSM715516) 67genomic small RNA (size selected RNA from t. (breast) | GSM339994(GSM339994) hues6. (cell line) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | SRR039191(GSM494810) PBMCs were isolated by ficoll gradient from t. (blood) | SRR095854(SRX039177) miRNA were isolated from FirstChoice Human Br. (brain) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR060982(GSM569186) Human centrocyte [09-001]. (cell line) | TAX577739(Rovira) total RNA. (breast) | SRR037937(GSM510475) 293cand2. (cell line) | SRR060169(GSM565979) 5-8F_cytoplasm. (cell line) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | TAX577745(Rovira) total RNA. (breast) | SRR039635(GSM518472) THP1_nuc_sRNAs. (cell line) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | SRR033730(GSM497075) Lymphoblastic (ALL411). (B cell) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033714(GSM497059) Burkitt Lymphoma (BL134). (B cell) | SRR033716(GSM497061) Mentle Cell Lymphoma (MCL114). (B cell) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | SRR189782 | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR039637(GSM518474) THP1_total_sRNAs. (cell line) | GSM450604(GSM450604) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577580(Rovira) total RNA. (breast) | SRR040015(GSM532900) G623T. (cervix) | SRR191571(GSM715681) 63genomic small RNA (size selected RNA from t. (breast) | SRR330859(SRX091697) tissue: skin psoriatic involveddisease state:. (skin) | DRR000560(DRX000318) Isolation of RNA following immunoprecipitatio. (ago1 cell line) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................CACAGACAGAGAGGGCTAAGGT......................................................................................... | 22 | 1 | 22.00 | 22.00 | - | 9.00 | - | - | 2.00 | - | 2.00 | 3.00 | - | - | 2.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - |
| ............................................................................................................................................ACAGACAGAGAGGGCTAAGGT......................................................................................... | 21 | 1 | 17.00 | 17.00 | 1.00 | - | 7.00 | - | - | 1.00 | 2.00 | - | - | 1.00 | - | - | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................CACAGACAGAGAGGGCTAAGGTT........................................................................................ | 23 | 1 | 9.00 | 22.00 | 1.00 | - | - | - | 1.00 | - | - | - | 3.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................ACAGACAGAGAGGGCTAAGGTG........................................................................................ | 22 | 1 | 5.00 | 5.00 | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................ACACAGACAGAGAGGGCTAAGGT......................................................................................... | 23 | 1 | 4.00 | 4.00 | 1.00 | - | - | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................CAGACAGAGAGGGCTAAGGTG........................................................................................ | 21 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................ACAGACAGAGAGGGCTAAGGTAA....................................................................................... | 23 | 1 | 4.00 | 17.00 | 1.00 | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................ACAGACAGAGAGGGCTAAGGTA........................................................................................ | 22 | 1 | 3.00 | 17.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................AGACAGAGAGGGCTAAGGTG........................................................................................ | 20 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................ACAGACAGAGAGGGCTAAGGA......................................................................................... | 21 | 1 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................CACAGACAGAGAGGGCTAA............................................................................................ | 19 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................AGACAGAGAGGGCTAAGGT......................................................................................... | 19 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................CACAGACAGAGAGGGCTAAGGTGT....................................................................................... | 24 | 1 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................ACAGACAGAGAGGGCTAAGGTGT....................................................................................... | 23 | 1 | 2.00 | 5.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................ACAGACAGAGAGGGCTAAGGTT........................................................................................ | 22 | 1 | 2.00 | 17.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................CACAGACAGAGAGGGCTAAGGG......................................................................................... | 22 | 2.00 | 0.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................CAGACAGAGAGGGCTAAGGTGCC...................................................................................... | 23 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................CACAGACAGAGAGGGCTAAGGGT........................................................................................ | 23 | 2.00 | 0.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................................AGCCCAGCCCTCTTCTGTCT......................................................... | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .............................................................................................................................................CAGACAGAGAGGGCTAAGGTGA....................................................................................... | 22 | 1 | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................ACAGACAGAGAGGGCTAAGACG........................................................................................ | 22 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................................AGCCCAGCCCTCTTCTGTCTCCTTT.................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................AGACAGAGAGGGCTAAGGTGTTT..................................................................................... | 23 | 1 | 1.00 | 3.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................AACCTCTGGGGAGGGCGGG......................................................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................ACACAGACAGAGAGGGCTAAGGATA....................................................................................... | 25 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................CAGACAGAGAGGGCTAAGG.......................................................................................... | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................GAGCTCTGGAGTCCCAGAAGC................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................CACAGACAGAGAGGGCTAAGGAG........................................................................................ | 23 | 1.00 | 0.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................AGACAGAGAGGGCTAAGGTGCCT..................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................GATCCGAGCTCTGGAGTCCCAGAAG.................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................CAGACAGAGAGGGCTAAGGTT........................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................CAGACAGAGAGGGCTAAGGT......................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................CAGACAGAGAGGGCTAAGGTAGAA..................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................CACAGACAGAGAGGGCTAAGGTG........................................................................................ | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................ACACAGACAGAGAGGGCTAAGA.......................................................................................... | 22 | 1.00 | 0.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................................AGCCCAGCCCTCTTCTGTCTCC....................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................ACAGACAGAGAGGGCTAAGAAT........................................................................................ | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................CAGACAGAGAGGGCTAAGGTGC....................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................CACAGACAGAGAGGGCAAAG........................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................ACAGACAGAGAGGGCTAAGG.......................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................CAGACAGAGAGGGCTAAGGTCA....................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................CAGACAGAGAGGGCTAAGGTA........................................................................................ | 21 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................ACACAGACAGAGAGGGCTAAGGG......................................................................................... | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................................................CCAGCCCTCTTCTGTCTCCCCT.................................................... | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | |
| ...........................................................................................................................................CACAGACAGAGAGGGCTAAGGA......................................................................................... | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................CAGACAGAGAGGGCTAAGGTAT....................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................ACAGACAGAGAGGGCTAAGGGA........................................................................................ | 22 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................CAGACAGAGAGGGCTAAGGTGAC...................................................................................... | 23 | 1 | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................CACAGACAGAGAGGGCTAAGGAGC....................................................................................... | 24 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................CACAGACAGAGAGGGCTCTGG.......................................................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................ACAGACAGAGAGGGCTAAGGTAGA...................................................................................... | 24 | 1 | 1.00 | 17.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................................................GAGCCCAGCCCTCTTCTGTCTCT....................................................... | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | |
| ..............................................................................................................................................AGACAGAGAGGGCTAAGGTAG....................................................................................... | 21 | 1 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................................................ACAGACAGAGAGGGCTAAGGTGA....................................................................................... | 23 | 1 | 1.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..........................................................................................................................................ACACAGACAGAGAGGGC............................................................................................... | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................TACACAGACAGAGAGGGCTAAGGTAT....................................................................................... | 26 | 1.00 | 0.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................ACAGACAGAGAGGGCTA............................................................................................. | 17 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................CAGACAGAGAGGGCTAAG........................................................................................... | 18 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - |
| TGCCCTTGTGGAGAAAGCAGTTTTCACTGCAACCTCTGGGGAGGGGAGCTCAACCCTGGGTCCCACTTTATCAAGAGTTTGGCTGCATGAGAATCACCCTGAAGCCCAGGCTTTCAACTACAGGGCTAGCCAGCCTGTACACAGACAGAGAGGGCTAAGGTGCCTGAGTGTGGAGCCCAGCCCTCTTCTGTCTCCCACAGTTTCAGATCCGAGCTCTGGAGTCCCAGAAGCGGCAGCAGGAGATGGTCCT ......................................................................................................................................((((....((((((((((((((..(((.((.......)).))).)))))))).))))))...)))).................................................. ......................................................................................................................................135..............................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | GSM339996(GSM339996) hues6Neuron. (cell line) | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450598(GSM450598) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450607(GSM450607) miRNA sequencing raw reads from post-mortem s. (brain) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | GSM450597(GSM450597) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039193(GSM494812) HL60 cell line is derived from acute promyelo. (cell line) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | GSM450599(GSM450599) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR060983(GSM569187) Human pre-germinal center B cell [09-001]. (cell line) | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR060985(GSM569189) Human naive B cell [09-001]. (cell line) | GSM450603(GSM450603) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039636(GSM518473) THP1_cyto_sRNAs. (cell line) | SRR191406(GSM715516) 67genomic small RNA (size selected RNA from t. (breast) | GSM339994(GSM339994) hues6. (cell line) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | SRR039191(GSM494810) PBMCs were isolated by ficoll gradient from t. (blood) | SRR095854(SRX039177) miRNA were isolated from FirstChoice Human Br. (brain) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR060982(GSM569186) Human centrocyte [09-001]. (cell line) | TAX577739(Rovira) total RNA. (breast) | SRR037937(GSM510475) 293cand2. (cell line) | SRR060169(GSM565979) 5-8F_cytoplasm. (cell line) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | TAX577745(Rovira) total RNA. (breast) | SRR039635(GSM518472) THP1_nuc_sRNAs. (cell line) | SRR094131(GSM651907) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | SRR033730(GSM497075) Lymphoblastic (ALL411). (B cell) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033714(GSM497059) Burkitt Lymphoma (BL134). (B cell) | SRR033716(GSM497061) Mentle Cell Lymphoma (MCL114). (B cell) | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | SRR189782 | SRR039622(GSM531985) HCV(+) Adjacent Tissue Sample. (liver) | SRR039637(GSM518474) THP1_total_sRNAs. (cell line) | GSM450604(GSM450604) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577580(Rovira) total RNA. (breast) | SRR040015(GSM532900) G623T. (cervix) | SRR191571(GSM715681) 63genomic small RNA (size selected RNA from t. (breast) | SRR330859(SRX091697) tissue: skin psoriatic involveddisease state:. (skin) | DRR000560(DRX000318) Isolation of RNA following immunoprecipitatio. (ago1 cell line) | SRR033722(GSM497067) KMS12 cell line (KMS12). (B cell) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................................................................AGTTTCAGATCCGAGCTG.................................. | 18 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................CAACTACAGGGCTAGCCGAGG................................................................................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................CCCAGGCTTTCAACTACGA............................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................GACAGAGAGGGCTAAGGTGCCTGAG.................................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................CTCAACCCTGGGTCCCA......................................................................................................................................................................................... | 17 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - |
| ..............................................................................................................................................................................................TCTCCCACAGTTTCAG............................................ | 16 | 5 | 0.20 | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.20 |