| (2) AGO1.ip | (1) AGO1.ip OTHER.mut | (3) AGO2.ip | (5) B-CELL | (5) BRAIN | (3) BREAST | (24) CELL-LINE | (1) CERVIX | (3) FIBROBLAST | (4) HEART | (2) HELA | (2) LIVER | (3) OTHER | (1) RRP40.ip | (10) SKIN | (1) UTERUS | (1) XRN.ip |
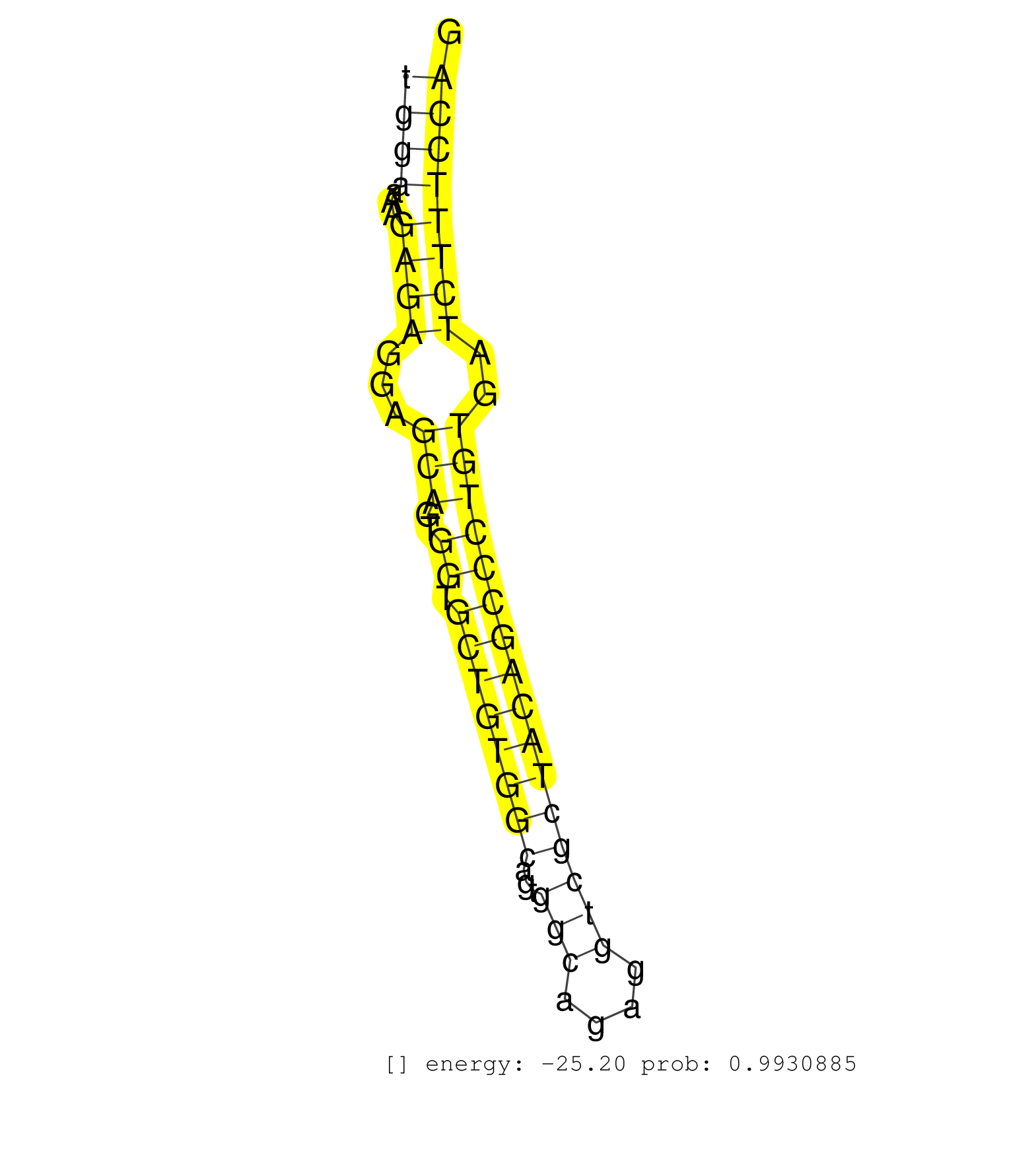
| ACAGGTCCTCCCCAGATACACCTGTGATCGGTGTTAATTCTGGCTTGAAAGAGCCCTTCAGAATGGGAAGTGTGGAAAGGTGCTAGGTTCTGATGGGTGGCATTGATTACATCCTGTCACTTAACCTCCTCCCTGGAAAAGAGAGGAGCAGTGGTGCTGTGGCAGTGGCAGAGGTCGCTACAGCCCTGTGATCTTTCCAGGAAAGAGACGGCTTTGGAGGCCTGGCAGCAGCCAGAGGAAGAGGTCGTGG .....................................................................................................................................((((...((((...(((..((.((((((((...(((....))))))))))))))))..))))))))................................................... .....................................................................................................................................134...............................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR189782 | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR033712(GSM497057) Burkitt Lymphoma (BL510). (B cell) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR207114(GSM721076) IP against AGO 1 & 2, RRP40 knockdown. (ago1/2 RRP40 cell line) | SRR363675(GSM830252) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | GSM1105749AGO2(GSM1105749) small RNA sequencing data. (ago2 hela) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | DRR000557(DRX000315) THP-1 whole cell RNA, after 3 day treatment w. (cell line) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | SRR029129(GSM416758) SW480. (cell line) | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR029126(GSM416755) 143B. (cell line) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | SRR037931(GSM510469) 293GFP. (cell line) | SRR033715(GSM497060) Mantle Cell Lymphoma (Mino122). (B cell) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR189786 | GSM416733(GSM416733) HEK293. (cell line) | SRR037933(GSM510471) 293cand4_rep2. (cell line) | DRR000559(DRX000317) THP-1 whole cell RNA, no treatment. (cell line) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | DRR000556(DRX000314) THP-1 whole cell RNA, after 3 day treatment w. (cell line) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR189784 | TAX577589(Rovira) total RNA. (breast) | SRR038855(GSM458538) D10. (cell line) | SRR033729(GSM497074) splenic MZL (Splenic414). (B cell) | GSM450603(GSM450603) miRNA sequencing raw reads from post-mortem s. (brain) | SRR037934(GSM510472) 293cand4_rep3. (cell line) | SRR330913(SRX091751) tissue: normal skindisease state: normal. (skin) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR040008(GSM532893) G727N. (cervix) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | SRR039193(GSM494812) HL60 cell line is derived from acute promyelo. (cell line) | GSM1105748AGO1(GSM1105748) small RNA sequencing data. (ago1 hela) | TAX577744(Rovira) total RNA. (breast) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR363676(GSM830253) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR326282(GSM769512) Dicer mRNA was knocked down using siDicer, to. (cell line) | GSM450599(GSM450599) miRNA sequencing raw reads from post-mortem s. (brain) | GSM359203(GSM359203) hepg2_untreated_b. (cell line) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330886(SRX091724) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR330899(SRX091737) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................................................................AAGAGAGGAGCAGTGGTGCTGTGG........................................................................................ | 24 | 1 | 17.00 | 17.00 | 3.00 | - | - | - | 1.00 | - | - | - | 2.00 | 2.00 | - | - | 3.00 | 1.00 | - | - | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................AGAGAGGAGCAGTGGTGCTGTGG........................................................................................ | 23 | 1 | 8.00 | 8.00 | - | - | - | - | - | - | - | 2.00 | - | 1.00 | - | - | - | - | - | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TACAGCCCTGTGATCTTTCCAGA................................................. | 23 | 1 | 7.00 | 6.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TCGCTACAGCCCTGTGATCTTTCCAG.................................................. | 26 | 1 | 6.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TACAGCCCTGTGATCTTTCCAG.................................................. | 22 | 1 | 6.00 | 6.00 | - | - | - | 1.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................AAGAGAGGAGCAGTGGTGCTGTGGA....................................................................................... | 25 | 1 | 4.00 | 17.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................AAGAGAGGAGCAGTGGTGCTGTAA........................................................................................ | 24 | 3.00 | 0.00 | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................CAGAATGGGAAGTGTGGAAAGGTG........................................................................................................................................................................ | 24 | 1 | 2.00 | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................ACAGCCCTGTGATCTTTCCA................................................... | 20 | 1 | 2.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TACAGCCCTGTGATCTTTCCA................................................... | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................AGGCCTGGCAGCAGCCAGAGGAAGAGGTCGTGG | 33 | 1 | 2.00 | 2.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................................................................................................GGCAGCAGCCAGAGGAAG......... | 18 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TACAGCCCTGTGATCTTTCC.................................................... | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................ACAGCCCTGTGATCTTTCCAG.................................................. | 21 | 1 | 2.00 | 2.00 | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................AGAGAGGAGCAGTGGTGCTGTGGTAT..................................................................................... | 26 | 1 | 2.00 | 8.00 | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TACAGCCCTGTGATCTTTCCATT................................................. | 23 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................................TGGAGGCCTGGCAGCAGCCAGAGGAAG......... | 27 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................AGGAAAGAGACGGCTTTGGAGG.............................. | 22 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................ACAGCCCTGTGATCTTTCCAGA................................................. | 22 | 1 | 1.00 | 2.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................TGGAAAAGAGAGGAGCAGTGGTGCT............................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................GGAAAGAGACGGCTTTGGAG............................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................CAGAATGGGAAGTGTGGCT............................................................................................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................AAGAGAGGAGCAGTGGTGCTGTGGTT...................................................................................... | 26 | 1 | 1.00 | 17.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................AAGAGAGGAGCAGTGGTGCTGCGGC....................................................................................... | 25 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................CTTGAAAGAGCCCTTCAGAA........................................................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................CTGGAAAAGAGAGGAGCAGT.................................................................................................. | 20 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................................AGACGGCTTTGGAGGCCTGGCAGCAGC.................. | 27 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TCGCTACAGCCCTGTGATCTTTCCAGA................................................. | 27 | 1 | 1.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................AGAGAGGAGCAGTGGTGCGT........................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................AAAGGTGCTAGGTTCTGCAG........................................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................................................................................................................................................................................TACAGCCCTGTGATCACT...................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................AGAGAGGAGCAGTGGTGCTGTGGC....................................................................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................AGCAGTGGTGCTGTGGAACG.................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..................CACCTGTGATCGGTGTTAATTCTGGC.............................................................................................................................................................................................................. | 26 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| ..............................................................ATGGGAAGTGTGGAAATTT......................................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................AAGAGAGGAGCAGTGGTGCTGTG......................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................AAAGAGCCCTTCAGAATGGGAAGTGTGGAAAGGTG........................................................................................................................................................................ | 35 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................AAGAGAGGAGCAGTGGTGCTGTGGC....................................................................................... | 25 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................................................AAGAGAGGAGCAGTGGTGCTGTGGT....................................................................................... | 25 | 1 | 1.00 | 17.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................AAAAGAGAGGAGCAGTGGTGCTGCGGC....................................................................................... | 27 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................................................................................................AAAGAGACGGCTTTGGAGG.............................. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................................................GGCTTTGGAGGCCTGGCAGCAGCCAGAGGAAG......... | 32 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................AGAGAGGAGCAGTGGTGCTGTGGA....................................................................................... | 24 | 1 | 1.00 | 8.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................AAAGAGACGGCTTTGGTAAG............................. | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................CAGAATGGGAAGTGTGGAT............................................................................................................................................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................................................................................GGAAAGAGACGGCTTTGGA................................ | 19 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................................AGAGGAAGAGGTCGTGA | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .........................................................................................................................................................................................................................AGGCCTGGCAGCAGCCAGAGGAAG......... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| .....................................................................................................................................................................................................CAGGAAAGAGACGGCTTTGGA................................ | 21 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................TCGCTACAGCCCTGTGATCTTA...................................................... | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ......................................................................................................................................................................................................AGGAAAGAGACGGCTTTGGA................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................TACAGCCCTGTGATCTTTCCAGT................................................. | 23 | 1 | 1.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................................................................AGGAAAGAGACGGCTTTGGAGGC............................. | 23 | 1 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................CGGCTTTGGAGGCCTGGCAGCAGCCAGAGG............ | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................................................................................................................AGAGGAAGAGGTCGTGG | 17 | 4 | 0.25 | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.25 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................................AGGAAGAGGTCGTGGTAT | 18 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - | - |
| ...........................................................................................................................................................................................................................................AGGAAGAGGTCGTGGGT | 17 | 6 | 0.17 | 0.17 | - | - | - | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................................................AGGAAGAGGTCGTGG | 15 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - |
| ................................................................................................................................CTCCCTGGAAAAGAGA.......................................................................................................... | 16 | 10 | 0.10 | 0.10 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.10 |
| ACAGGTCCTCCCCAGATACACCTGTGATCGGTGTTAATTCTGGCTTGAAAGAGCCCTTCAGAATGGGAAGTGTGGAAAGGTGCTAGGTTCTGATGGGTGGCATTGATTACATCCTGTCACTTAACCTCCTCCCTGGAAAAGAGAGGAGCAGTGGTGCTGTGGCAGTGGCAGAGGTCGCTACAGCCCTGTGATCTTTCCAGGAAAGAGACGGCTTTGGAGGCCTGGCAGCAGCCAGAGGAAGAGGTCGTGG .....................................................................................................................................((((...((((...(((..((.((((((((...(((....))))))))))))))))..))))))))................................................... .....................................................................................................................................134...............................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR039623(GSM531986) HCV(+) HCC Tissue Sample. (liver) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR189782 | SRR060168(GSM565978) 5-8F_nucleus. (cell line) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR033712(GSM497057) Burkitt Lymphoma (BL510). (B cell) | SRR326279(GSM769509) cytoplasmic fraction was isolated using PARIS. (cell line) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR207114(GSM721076) IP against AGO 1 & 2, RRP40 knockdown. (ago1/2 RRP40 cell line) | SRR363675(GSM830252) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | GSM1105749AGO2(GSM1105749) small RNA sequencing data. (ago2 hela) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | DRR000557(DRX000315) THP-1 whole cell RNA, after 3 day treatment w. (cell line) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | SRR029129(GSM416758) SW480. (cell line) | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | SRR029126(GSM416755) 143B. (cell line) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | SRR037931(GSM510469) 293GFP. (cell line) | SRR033715(GSM497060) Mantle Cell Lymphoma (Mino122). (B cell) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR189786 | GSM416733(GSM416733) HEK293. (cell line) | SRR037933(GSM510471) 293cand4_rep2. (cell line) | DRR000559(DRX000317) THP-1 whole cell RNA, no treatment. (cell line) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR037876(GSM522374) fibroblasts_cell_culture. (fibroblast) | DRR000556(DRX000314) THP-1 whole cell RNA, after 3 day treatment w. (cell line) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR189784 | TAX577589(Rovira) total RNA. (breast) | SRR038855(GSM458538) D10. (cell line) | SRR033729(GSM497074) splenic MZL (Splenic414). (B cell) | GSM450603(GSM450603) miRNA sequencing raw reads from post-mortem s. (brain) | SRR037934(GSM510472) 293cand4_rep3. (cell line) | SRR330913(SRX091751) tissue: normal skindisease state: normal. (skin) | SRR094130(GSM651906) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR040008(GSM532893) G727N. (cervix) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | SRR039193(GSM494812) HL60 cell line is derived from acute promyelo. (cell line) | GSM1105748AGO1(GSM1105748) small RNA sequencing data. (ago1 hela) | TAX577744(Rovira) total RNA. (breast) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | SRR363676(GSM830253) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR326282(GSM769512) Dicer mRNA was knocked down using siDicer, to. (cell line) | GSM450599(GSM450599) miRNA sequencing raw reads from post-mortem s. (brain) | GSM359203(GSM359203) hepg2_untreated_b. (cell line) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR330887(SRX091725) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330886(SRX091724) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR033713(GSM497058) Burkitt Lymphoma (BL115). (B cell) | SRR330899(SRX091737) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................................CGCTACAGCCCTGTG............................................................ | 15 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................TGAAAGAGCCCTTCA.............................................................................................................................................................................................. | 15 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - | - | - |
| ..............................................GAAAGAGCCCTTCAG............................................................................................................................................................................................. | 15 | 9 | 0.11 | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 | - |