| (1) AGO1.ip | (3) AGO2.ip | (1) AGO3.ip | (2) B-CELL | (11) BRAIN | (5) BREAST | (13) CELL-LINE | (3) CERVIX | (9) HEART | (4) HELA | (1) KIDNEY | (2) LIVER | (3) OTHER | (1) OTHER.ip | (16) SKIN | (1) XRN.ip |
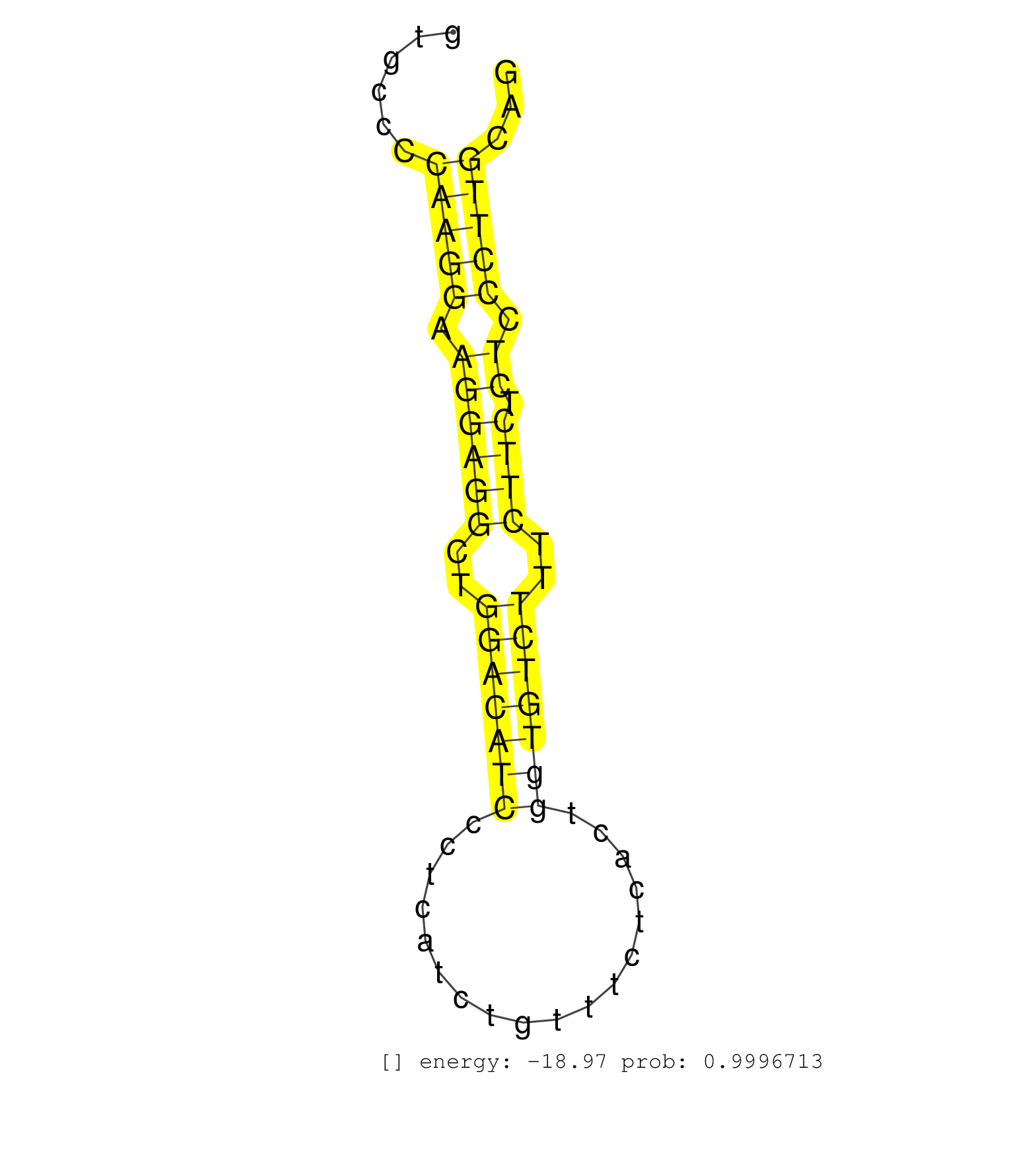
| CCTCTTTCTTCCTGAAGTTGCGAGATAGAGCCCCATGAGTGCCTAGGCCCCCTTTAACTCCAAGTCCCCATAATCCTCAGAGAGCTGACATGTTCTTATCCCAGGGGACTTGCTTCTGTGCTGGTATTCTGTGCCCCAAGGAAGGAGGCTGGACATCCCTCATCTGTTTCTCACTGGTGTCTTTCTTCTCTCCCTTGCAGGGCACATGACTGACCTGATTTATGCAGAGAAAGAGCTGGTGCAGTCTCTG ........................................................................................................................................(((((.((((((..(((((((..................)))))))..)))).)).)))))..................................................... ..................................................................................................................................131..................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | GSM1105748AGO1(GSM1105748) small RNA sequencing data. (ago1 hela) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | GSM450599(GSM450599) miRNA sequencing raw reads from post-mortem s. (brain) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039614(GSM531977) HBV-infected Liver Tissue. (liver) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | SRR553574(SRX182780) source: Heart. (Heart) | SRR029125(GSM416754) U2OS. (cell line) | SRR033711(GSM497056) GCB DLBCL (GCB110). (B cell) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450607(GSM450607) miRNA sequencing raw reads from post-mortem s. (brain) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR038852(GSM458535) QF1160MB. (cell line) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | GSM1105750AGO3(GSM1105750) small RNA sequencing data. (ago3 hela) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | GSM1105749AGO2(GSM1105749) small RNA sequencing data. (ago2 hela) | SRR330886(SRX091724) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR390723(GSM850202) total small RNA. (cell line) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | SRR191602(GSM715712) 60genomic small RNA (size selected RNA from t. (breast) | SRR038854(GSM458537) MM653. (cell line) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR330869(SRX091707) tissue: skin psoriatic involveddisease state:. (skin) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | DRR000561(DRX000319) Isolation of RNA following immunoprecipitatio. (ago2 cell line) | GSM450603(GSM450603) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330862(SRX091700) tissue: skin psoriatic involveddisease state:. (skin) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | TAX577741(Rovira) total RNA. (breast) | GSM532876(GSM532876) G547T. (cervix) | SRR326281(GSM769511) Dicer mRNA was knocked down using siDicer, cy. (cell line) | SRR330888(SRX091726) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577745(Rovira) total RNA. (breast) | SRR189786 | SRR330912(SRX091750) tissue: normal skindisease state: normal. (skin) | SRR330894(SRX091732) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM1105751AGO4(GSM1105751) small RNA sequencing data. (ago4 hela) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR040028(GSM532913) G026N. (cervix) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | SRR553575(SRX182781) source: Kidney. (Kidney) | SRR191604(GSM715714) 74genomic small RNA (size selected RNA from t. (breast) | SRR330865(SRX091703) tissue: skin psoriatic involveddisease state:. (skin) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | GSM450598(GSM450598) miRNA sequencing raw reads from post-mortem s. (brain) | SRR040017(GSM532902) G645T. (cervix) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) | SRR444059(SRX128907) Sample 17cDNABarcode: AF-PP-339: ACG CTC TTC . (skin) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................................CCAAGGAAGGAGGCTGGACATC............................................................................................. | 22 | 1 | 23.00 | 23.00 | 3.00 | 3.00 | - | 2.00 | - | - | - | 1.00 | 2.00 | - | - | - | 1.00 | 2.00 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TGTCTTTCTTCTCTCCCTTGCAG.................................................. | 23 | 1 | 19.00 | 19.00 | - | 1.00 | 2.00 | - | - | 2.00 | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | 1.00 | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - |
| .......................................................................................................................................CCAAGGAAGGAGGCTGGACAT.............................................................................................. | 21 | 1 | 18.00 | 18.00 | 2.00 | - | - | 3.00 | 3.00 | - | 1.00 | - | - | - | 2.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ......................................................................................................................................CCCAAGGAAGGAGGCTGGACAT.............................................................................................. | 22 | 1 | 8.00 | 8.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | 1.00 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................CCCAAGGAAGGAGGCTGGACATC............................................................................................. | 23 | 1 | 7.00 | 7.00 | - | 1.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................CCCAAGGAAGGAGGCTGGACA............................................................................................... | 21 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TGTCTTTCTTCTCTCCCTTGC.................................................... | 21 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TGTCTTTCTTCTCTCCCTTGCA................................................... | 22 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TCTTTCTTCTCTCCCTTGCAGT................................................. | 22 | 1 | 3.00 | 1.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................CCAAGGAAGGAGGCTGGACATCA............................................................................................ | 23 | 1 | 3.00 | 23.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................CCCAAGGAAGGAGGCTGGACATCA............................................................................................ | 24 | 1 | 2.00 | 7.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................CCCCAAGGAAGGAGGCTGGACATC............................................................................................. | 24 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................CCAAGGAAGGAGGCTGGACA............................................................................................... | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................................................................................TGACCTGATTTATGCAGACAA................... | 21 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................CCCCAAGGAAGGAGGCTGGACA............................................................................................... | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................CCAAGGAAGGAGGCTGGACATCT............................................................................................ | 23 | 1 | 2.00 | 23.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - |
| .......................................................................................................................................CCAAGGAAGGAGGCTGGACATT............................................................................................. | 22 | 1 | 2.00 | 18.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................CCAAGGAAGGAGGCTGGACAA.............................................................................................. | 21 | 1 | 1.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................................................................................................................ACATGACTGACCTGATTTATGCAGAGAAAG................. | 30 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................TCTTTCTTCTCTCCCTTGCAG.................................................. | 21 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................CCAAGGAAGGAGGCTGGACACA............................................................................................. | 22 | 1 | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................CCCAAGGAAGGAGGCTGGTCAT.............................................................................................. | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................................................................................................................................................................................CACATGACTGACCTGATTTATGCAGA...................... | 26 | 1 | 1.00 | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TGTCTTTCTTCTCTCCCTTGCAGT................................................. | 24 | 1 | 1.00 | 19.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..................................................................................................................................................GGCTGGACATCCCTCATCTGT................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................................................................................TGACTGACCTGATTTATGCAGAGAAAGAGCTGGT.......... | 34 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................................................................................ACTGACCTGATTTATGC......................... | 17 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................AAGGAAGGAGGCTGGACTTAC............................................................................................ | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........CCTGAAGTTGCGAGATAGAGC........................................................................................................................................................................................................................... | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .................................................................................................................................................................................TGTCTTTCTTCTCTCCCGCAG.................................................... | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................CCAAGGAAGGAGGCTGGACATA............................................................................................. | 22 | 1 | 1.00 | 18.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................CAAGGAAGGAGGCTGGACATA............................................................................................. | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................TCTTTCTTCTCTCCCTTGCAGTAG............................................... | 24 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................................................................GGGCACATGACTGACCTGATTTATGCAGAGAA................... | 32 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................CCAAGGAAGGAGGCTGGACATAGA........................................................................................... | 24 | 1 | 1.00 | 18.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................................................TGTCTTTCTTCTCTCCCTTACA................................................... | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................CAAGGAAGGAGGCTGGACA............................................................................................... | 19 | 2 | 0.50 | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .........................................................................................................................................AAGGAAGGAGGCTGGACAT.............................................................................................. | 19 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - |
| ...................................................................................................................CTGTGCTGGTATTCT........................................................................................................................ | 15 | 6 | 0.17 | 0.17 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.17 | - |
| .................................CATGAGTGCCTAGG........................................................................................................................................................................................................... | 14 | 9 | 0.11 | 0.11 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.11 |
| CCTCTTTCTTCCTGAAGTTGCGAGATAGAGCCCCATGAGTGCCTAGGCCCCCTTTAACTCCAAGTCCCCATAATCCTCAGAGAGCTGACATGTTCTTATCCCAGGGGACTTGCTTCTGTGCTGGTATTCTGTGCCCCAAGGAAGGAGGCTGGACATCCCTCATCTGTTTCTCACTGGTGTCTTTCTTCTCTCCCTTGCAGGGCACATGACTGACCTGATTTATGCAGAGAAAGAGCTGGTGCAGTCTCTG ........................................................................................................................................(((((.((((((..(((((((..................)))))))..)))).)).)))))..................................................... ..................................................................................................................................131..................................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | GSM1105748AGO1(GSM1105748) small RNA sequencing data. (ago1 hela) | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR342895(SRX096791) small RNA seq of Left atrial tissue. (heart) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | GSM450599(GSM450599) miRNA sequencing raw reads from post-mortem s. (brain) | SRR342898(SRX096794) small RNA seq of Right atrial tissue. (heart) | SRR342900(SRX096796) small RNA seq of Right atrial tissue. (heart) | SRR033724(GSM497069) L428 cell line (L428). (B cell) | SRR029054(GSM402329) MCF7_smallRNAseq. (cell line) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | SRR342894(SRX096790) small RNA seq of Right atrial tissue. (heart) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR342901(SRX096797) small RNA seq of Left atrial tissue. (heart) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | SRR039614(GSM531977) HBV-infected Liver Tissue. (liver) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | SRR553574(SRX182780) source: Heart. (Heart) | SRR029125(GSM416754) U2OS. (cell line) | SRR033711(GSM497056) GCB DLBCL (GCB110). (B cell) | GSM450602(GSM450602) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450607(GSM450607) miRNA sequencing raw reads from post-mortem s. (brain) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR038852(GSM458535) QF1160MB. (cell line) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | GSM1105750AGO3(GSM1105750) small RNA sequencing data. (ago3 hela) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | GSM1105749AGO2(GSM1105749) small RNA sequencing data. (ago2 hela) | SRR330886(SRX091724) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR390723(GSM850202) total small RNA. (cell line) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | SRR191602(GSM715712) 60genomic small RNA (size selected RNA from t. (breast) | SRR038854(GSM458537) MM653. (cell line) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR330869(SRX091707) tissue: skin psoriatic involveddisease state:. (skin) | SRR342897(SRX096793) small RNA seq of Left atrial tissue. (heart) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | DRR000561(DRX000319) Isolation of RNA following immunoprecipitatio. (ago2 cell line) | GSM450603(GSM450603) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330862(SRX091700) tissue: skin psoriatic involveddisease state:. (skin) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | TAX577741(Rovira) total RNA. (breast) | GSM532876(GSM532876) G547T. (cervix) | SRR326281(GSM769511) Dicer mRNA was knocked down using siDicer, cy. (cell line) | SRR330888(SRX091726) tissue: skin psoriatic uninvolveddisease stat. (skin) | TAX577745(Rovira) total RNA. (breast) | SRR189786 | SRR330912(SRX091750) tissue: normal skindisease state: normal. (skin) | SRR330894(SRX091732) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM1105751AGO4(GSM1105751) small RNA sequencing data. (ago4 hela) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR040028(GSM532913) G026N. (cervix) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | SRR553575(SRX182781) source: Kidney. (Kidney) | SRR191604(GSM715714) 74genomic small RNA (size selected RNA from t. (breast) | SRR330865(SRX091703) tissue: skin psoriatic involveddisease state:. (skin) | SRR039192(GSM494811) K562 cell line is derived from a CML patient . (cell line) | GSM450598(GSM450598) miRNA sequencing raw reads from post-mortem s. (brain) | SRR040017(GSM532902) G645T. (cervix) | SRR326280(GSM769510) total cell content of unperturbed cells was s. (cell line) | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) | SRR444059(SRX128907) Sample 17cDNABarcode: AF-PP-339: ACG CTC TTC . (skin) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................................................................................GACCTGATTTATGCATACA.................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................CCCCATAATCCTCAGATAGG..................................................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................................CTGGTGTCTTTCTTCAGCA.......................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |