| (1) AGO2.ip | (9) BRAIN | (8) BREAST | (7) CELL-LINE | (2) HEART | (2) LIVER | (3) OTHER | (17) SKIN |
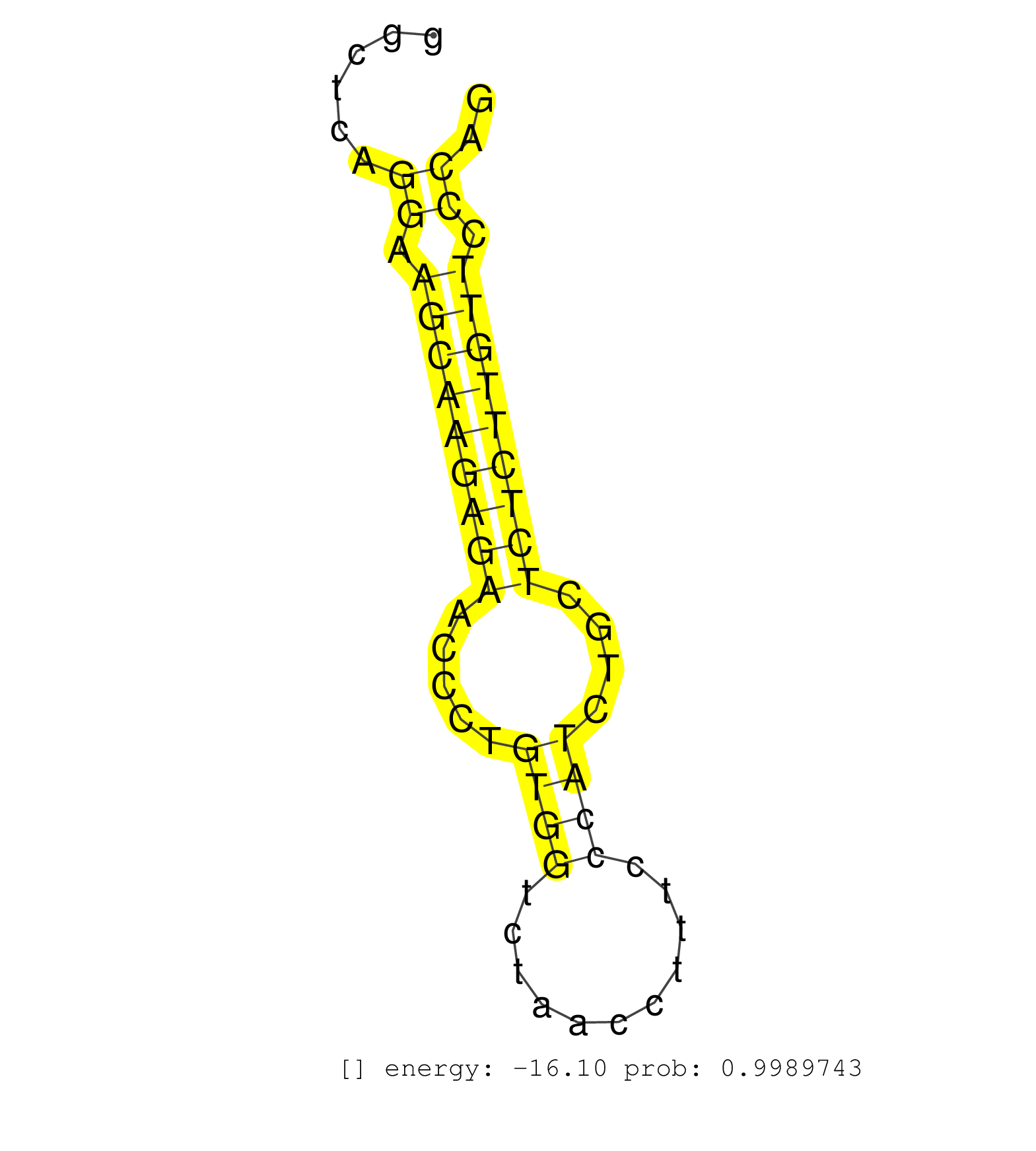
| TCTCCAGCCCCCTTTTTTCTTGCTGAGTTGTTTTAAAAGTTAAAAAGTGCTTAGTGACCTTGGACAACATTAGCACCTTCTAAGTCCTGGAATCCACCAAGAAGGAGGAAGGGAGATGGTGCTGAATTCCCCTAAGGCAGGGCTCAGGAAGCAAGAGAACCCTGTGGTCTAACCTTTCCCATCTGCTCTCTTGTTCCCAGGCTACGTGCAGGCCTGCCGCGCCCTGATGATTGCTGCCTCGGTCCTGGGT ..................................................................................................................................................((.(((((((((.....((((...........))))....))))))))).)).................................................... ............................................................................................................................................141........................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | GSM450603(GSM450603) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189784 | SRR029129(GSM416758) SW480. (cell line) | SRR330899(SRX091737) tissue: skin psoriatic uninvolveddisease stat. (skin) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | TAX577745(Rovira) total RNA. (breast) | SRR189785 | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR029126(GSM416755) 143B. (cell line) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR330872(SRX091710) tissue: skin psoriatic involveddisease state:. (skin) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330885(SRX091723) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM450604(GSM450604) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577580(Rovira) total RNA. (breast) | TAX577743(Rovira) total RNA. (breast) | SRR343334 | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) | SRR039612(GSM531975) Human Normal Liver Tissue Sample 2. (liver) | SRR191547(GSM715657) 155genomic small RNA (size selected RNA from . (breast) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | SRR343335 | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | SRR330886(SRX091724) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330875(SRX091713) tissue: skin psoriatic involveddisease state:. (skin) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | TAX577588(Rovira) total RNA. (breast) | SRR330891(SRX091729) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR189787 | SRR095854(SRX039177) miRNA were isolated from FirstChoice Human Br. (brain) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR037937(GSM510475) 293cand2. (cell line) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) | SRR330871(SRX091709) tissue: skin psoriatic involveddisease state:. (skin) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577590(Rovira) total RNA. (breast) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | SRR191554(GSM715664) 99genomic small RNA (size selected RNA from t. (breast) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................AGGAAGCAAGAGAACCCTGTGG................................................................................... | 22 | 1 | 30.00 | 30.00 | 6.00 | 2.00 | 3.00 | 1.00 | - | 1.00 | 1.00 | - | - | - | 2.00 | - | - | - | - | 2.00 | 1.00 | 1.00 | - | - | 1.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | 1.00 | - | 1.00 | 1.00 |
| ...............................................................................................................................................TCAGGAAGCAAGAGAACCCTGT..................................................................................... | 22 | 1 | 10.00 | 10.00 | 2.00 | 3.00 | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| .................................................................................................................................................AGGAAGCAAGAGAACCCTGT..................................................................................... | 20 | 1 | 7.00 | 7.00 | 2.00 | 2.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................ATCTGCTCTCTTGTTCCCAGT................................................. | 21 | 5.00 | 0.00 | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................AGGAAGCAAGAGAACCCTGTGGA.................................................................................. | 23 | 1 | 4.00 | 30.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................TCAGGAAGCAAGAGAACCCTGTGG................................................................................... | 24 | 1 | 4.00 | 4.00 | 1.00 | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - |
| ...............................................................................................................................................TCAGGAAGCAAGAGAACCCT....................................................................................... | 20 | 1 | 4.00 | 4.00 | - | 2.00 | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................................................TCAGGAAGCAAGAGAACCCTGTG.................................................................................... | 23 | 1 | 3.00 | 3.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................AGGAAGCAAGAGAACCCTGTGA................................................................................... | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................AGGAAGCAAGAGAACCCTGTG.................................................................................... | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................................................................CATCTGCTCTCTTGTTCCCAGT................................................. | 22 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................ATCTGCTCTCTTGTTCCCAGTA................................................ | 22 | 2.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................................................CTAACCTTTCCCATCACAG............................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................................................................................................GCCGCGCCCTGATGATTGCTGCCTCGGTCCT.... | 31 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................AGGAAGCAAGAGAACCCTGTGGTTTA............................................................................... | 26 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................AGGGAGATGGTGCTGAC............................................................................................................................ | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................TTGCTGAGTTGTTTTAACAA................................................................................................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................TCAGGAAGCAAGAGAACCCTGTGA................................................................................... | 24 | 1 | 1.00 | 3.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................ATCTGCTCTCTTGTTCCCAGTAT............................................... | 23 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................................................................................AGGAAGCAAGAGAACCG........................................................................................ | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................TCAGGAAGCAAGAGAACCCTGA..................................................................................... | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .....................................................................................................................................................................................................................................ATTGCTGCCTCGGTCTGA... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................................................GGGCTCAGGAAGCAATAG............................................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................CAGGAAGCAAGAGAACCCTGTGG................................................................................... | 23 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................AGGAAGCAAGAGAACCCTGTGGTCT................................................................................ | 25 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................AGAAGGAGGAAGGGAGGCG.................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................AAAAGTGCTTAGTGACCTTG............................................................................................................................................................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................AGGAAGCAAGAGAACCCTGTGAA.................................................................................. | 23 | 1 | 1.00 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................AGGAAGCAAGAGAACCCTGTGGAA................................................................................. | 24 | 1 | 1.00 | 30.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................................................CTCAGGAAGCAAGAGAACCCTG...................................................................................... | 22 | 1 | 1.00 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ................................................................................................................................................CAGGAAGCAAGAGAACCCTGTG.................................................................................... | 22 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ....................................................................................................................................................................................ATCTGCTCTCTTGTTCCCAGTT................................................ | 22 | 1.00 | 0.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ................................................................................................................................................CAGGAAGCAAGAGAACCC........................................................................................ | 18 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .................................................................................................................................................AGGAAGCAAGAGAACCCTGTGGTGT................................................................................ | 25 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...............................................................................................................................................TCAGGAAGCAAGAGAACCCTAA..................................................................................... | 22 | 1 | 1.00 | 4.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| TCTCCAGCCCCCTTTTTTCTTGCTGAGTTGTTTTAAAAGTTAAAAAGTGCTTAGTGACCTTGGACAACATTAGCACCTTCTAAGTCCTGGAATCCACCAAGAAGGAGGAAGGGAGATGGTGCTGAATTCCCCTAAGGCAGGGCTCAGGAAGCAAGAGAACCCTGTGGTCTAACCTTTCCCATCTGCTCTCTTGTTCCCAGGCTACGTGCAGGCCTGCCGCGCCCTGATGATTGCTGCCTCGGTCCTGGGT ..................................................................................................................................................((.(((((((((.....((((...........))))....))))))))).)).................................................... ............................................................................................................................................141........................................................200................................................ | Size | Perfect hit | Total Norm | Perfect Norm | GSM450603(GSM450603) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450601(GSM450601) miRNA sequencing raw reads from post-mortem s. (brain) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | GSM450610(GSM450610) miRNA sequencing raw reads from post-mortem s. (brain) | GSM450600(GSM450600) miRNA sequencing raw reads from post-mortem s. (brain) | SRR189779(GSM714639) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR189784 | SRR029129(GSM416758) SW480. (cell line) | SRR330899(SRX091737) tissue: skin psoriatic uninvolveddisease stat. (skin) | RoviraIPAgo2(Rovira) total RNA. (ago2 breast) | TAX577745(Rovira) total RNA. (breast) | SRR189785 | GSM450609(GSM450609) miRNA sequencing raw reads from post-mortem s. (brain) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR029126(GSM416755) 143B. (cell line) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR330872(SRX091710) tissue: skin psoriatic involveddisease state:. (skin) | SRR189781(GSM714641) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330885(SRX091723) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM450604(GSM450604) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577580(Rovira) total RNA. (breast) | TAX577743(Rovira) total RNA. (breast) | SRR343334 | SRR342899(SRX096795) small RNA seq of Left atrial tissue. (heart) | SRR330905(SRX091743) tissue: normal skindisease state: normal. (skin) | SRR039612(GSM531975) Human Normal Liver Tissue Sample 2. (liver) | SRR191547(GSM715657) 155genomic small RNA (size selected RNA from . (breast) | SRR039190(GSM494809) PBMCs were isolated by ficoll gradient from t. (blood) | SRR343335 | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | SRR330886(SRX091724) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330875(SRX091713) tissue: skin psoriatic involveddisease state:. (skin) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | TAX577588(Rovira) total RNA. (breast) | SRR330891(SRX091729) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR189787 | SRR095854(SRX039177) miRNA were isolated from FirstChoice Human Br. (brain) | SRR553573(SRX182779) source: Cerebellum. (Cerebellum) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR037937(GSM510475) 293cand2. (cell line) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) | SRR330871(SRX091709) tissue: skin psoriatic involveddisease state:. (skin) | SRR330879(SRX091717) tissue: skin psoriatic involveddisease state:. (skin) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577590(Rovira) total RNA. (breast) | SRR107297(GSM677704) 18-30nt fraction of small RNA. (cell line) | SRR191554(GSM715664) 99genomic small RNA (size selected RNA from t. (breast) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................CTTCTAAGTCCTGGAATCTG.......................................................................................................................................................... | 20 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................GAAGGGAGATGGTGCTCT............................................................................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................GTCCTGGAATCCACCAACATG.................................................................................................................................................. | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .............................................................................................................................................................................CTTTCCCATCTGCTCTTTG.......................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................AAGCAAGAGAACCCTGTAT................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .................................................................................AAGTCCTGGAATCCACCAATTT................................................................................................................................................... | 22 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | |
| ...........................................................................................................GAAGGGAGATGGTGCTCTT............................................................................................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ...................................................................................................................................................................................CATCTGCTCTCTTGTCTGG.................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................................................................................................................................................................CCAGGCTACGTGCAGAGGT................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..........................................AAAAGTGCTTAGTGAACG.............................................................................................................................................................................................. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................................................................................................................................TGCTGCCTCGGTCCTGTT. | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ........................................................................................................................................................................................................................................GCTGCCTCGGTCCTGGG. | 17 | 3 | 0.33 | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................AAGGAGGAAGGGAGATG.................................................................................................................................... | 17 | 7 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |