| (1) AGO1.ip | (1) AGO1.ip OTHER.mut | (4) AGO2.ip | (7) B-CELL | (5) BRAIN | (20) BREAST | (27) CELL-LINE | (7) CERVIX | (2) FIBROBLAST | (1) HEART | (1) HELA | (12) LIVER | (1) OTHER | (2) RRP40.ip | (49) SKIN | (1) UTERUS | (1) XRN.ip |
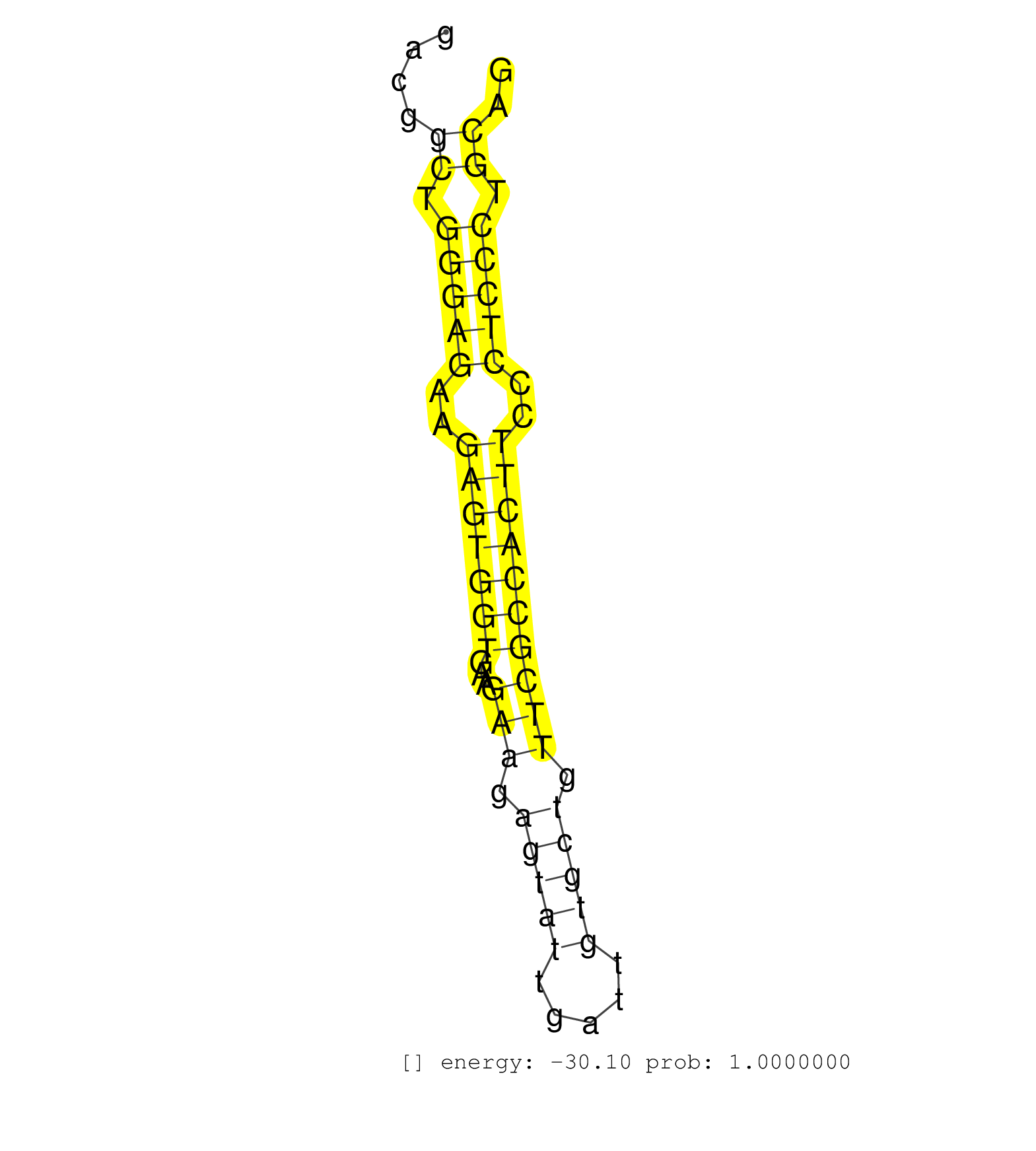
| CACCTTCCAGTATTTCATCTGCATTGGCATCCCACCTGCTCCTTGCCGAGGTGAGCCTGCCCTTCCCTTGAGCCGTCTACTTTTAGAAGAAGCCATTACTGAGCGGACGGCTGGGAGAAGAGTGGTGAAGAAGAGTATTGATTGTGCTGTTCGCCACTTCCCTCCCTGCAGATTACCCGTGGAGATTCAAGGGTGCCAGCTTCAATGACAACATCATAAAG .............................................................................................................((.(((((..(((((((...(((.(((((.....))))).))))))))))..))))).)).................................................... .........................................................................................................106..............................................................171................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR207114(GSM721076) IP against AGO 1 & 2, RRP40 knockdown. (ago1/2 RRP40 cell line) | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR033716(GSM497061) Mentle Cell Lymphoma (MCL114). (B cell) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR039614(GSM531977) HBV-infected Liver Tissue. (liver) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR029124(GSM416753) HeLa. (hela) | SRR033728(GSM497073) MALT (MALT413). (B cell) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR039612(GSM531975) Human Normal Liver Tissue Sample 2. (liver) | SRR039611(GSM531974) Human Normal Liver Tissue Sample 1. (liver) | SRR033729(GSM497074) splenic MZL (Splenic414). (B cell) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | SRR330894(SRX091732) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR040035(GSM532920) G001T. (cervix) | SRR330895(SRX091733) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330865(SRX091703) tissue: skin psoriatic involveddisease state:. (skin) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR040014(GSM532899) G623N. (cervix) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | SRR033712(GSM497057) Burkitt Lymphoma (BL510). (B cell) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR444057(SRX128905) Sample 15cDNABarcode: AF-PP-334: ACG CTC TTC . (skin) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330903(SRX091741) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | SRR330904(SRX091742) tissue: normal skindisease state: normal. (skin) | SRR207118(GSM721080) RRP40 knockdown. (RRP40 cell line) | TAX577579(Rovira) total RNA. (breast) | SRR330900(SRX091738) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR038861(GSM458544) MM466. (cell line) | TAX577739(Rovira) total RNA. (breast) | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | SRR363675(GSM830252) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR330913(SRX091751) tissue: normal skindisease state: normal. (skin) | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR330898(SRX091736) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR330891(SRX091729) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR038856(GSM458539) D11. (cell line) | SRR330923(SRX091761) tissue: normal skindisease state: normal. (skin) | SRR343333(GSM796036) KSHV (HHV8). (cell line) | TAX577744(Rovira) total RNA. (breast) | SRR039624(GSM531987) HBV(-) HCV(-) Adjacent Tissue Sample. (liver) | TAX577746(Rovira) total RNA. (breast) | TAX577589(Rovira) total RNA. (breast) | SRR189782 | SRR038855(GSM458538) D10. (cell line) | SRR330912(SRX091750) tissue: normal skindisease state: normal. (skin) | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | SRR330880(SRX091718) tissue: skin psoriatic involveddisease state:. (skin) | SRR330875(SRX091713) tissue: skin psoriatic involveddisease state:. (skin) | TAX577741(Rovira) total RNA. (breast) | SRR330874(SRX091712) tissue: skin psoriatic involveddisease state:. (skin) | SRR444053(SRX128901) Sample 13cDNABarcode: AF-PP-341: ACG CTC TTC . (skin) | GSM956925AGO2Paz8(GSM956925) cell line: HEK293 cell linepassages: 15-20ip . (ago2 cell line) | SRR444059(SRX128907) Sample 17cDNABarcode: AF-PP-339: ACG CTC TTC . (skin) | SRR330916(SRX091754) tissue: normal skindisease state: normal. (skin) | SRR033730(GSM497075) Lymphoblastic (ALL411). (B cell) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR191549(GSM715659) 105genomic small RNA (size selected RNA from . (breast) | GSM450599(GSM450599) miRNA sequencing raw reads from post-mortem s. (brain) | SRR363673(GSM830250) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR191500(GSM715610) 7genomic small RNA (size selected RNA from to. (breast) | TAX577588(Rovira) total RNA. (breast) | SRR330870(SRX091708) tissue: skin psoriatic involveddisease state:. (skin) | SRR191418(GSM715528) 40genomic small RNA (size selected RNA from t. (breast) | SRR330897(SRX091735) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191419(GSM715529) 41genomic small RNA (size selected RNA from t. (breast) | SRR038863(GSM458546) MM603. (cell line) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577742(Rovira) total RNA. (breast) | SRR343336 | SRR040010(GSM532895) G529N. (cervix) | SRR330914(SRX091752) tissue: normal skindisease state: normal. (skin) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | DRR000561(DRX000319) Isolation of RNA following immunoprecipitatio. (ago2 cell line) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | GSM956925Ago2PAZ(GSM956925) cell line: HEK293 cell linepassages: 15-20ip . (ago2 cell line) | SRR038854(GSM458537) MM653. (cell line) | SRR191551(GSM715661) 32genomic small RNA (size selected RNA from t. (breast) | SRR191602(GSM715712) 60genomic small RNA (size selected RNA from t. (breast) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | SRR343337 | GSM359198(GSM359198) hepg2_cip_tap_pasr_a. (cell line) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | SRR330892(SRX091730) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | SRR191625(GSM715735) 32genomic small RNA (size selected RNA from t. (breast) | SRR039637(GSM518474) THP1_total_sRNAs. (cell line) | SRR040017(GSM532902) G645T. (cervix) | SRR040036(GSM532921) G243N. (cervix) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR191578(GSM715688) 100genomic small RNA (size selected RNA from . (breast) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR330918(SRX091756) tissue: normal skindisease state: normal. (skin) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | GSM359203(GSM359203) hepg2_untreated_b. (cell line) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR191605(GSM715715) 76genomic small RNA (size selected RNA from t. (breast) | SRR038862(GSM458545) MM472. (cell line) | SRR330911(SRX091749) tissue: normal skindisease state: normal. (skin) | SRR191554(GSM715664) 99genomic small RNA (size selected RNA from t. (breast) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR330881(SRX091719) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191580(GSM715690) 103genomic small RNA (size selected RNA from . (breast) | GSM450603(GSM450603) miRNA sequencing raw reads from post-mortem s. (brain) | SRR189777(GSM714637) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR191603(GSM715713) 71genomic small RNA (size selected RNA from t. (breast) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | DRR000556(DRX000314) THP-1 whole cell RNA, after 3 day treatment w. (cell line) | SRR330860(SRX091698) tissue: skin psoriatic involveddisease state:. (skin) | SRR040015(GSM532900) G623T. (cervix) | SRR040027(GSM532912) G220T. (cervix) | SRR444052(SRX128900) Sample 12cDNABarcode: AF-PP-340: ACG CTC TTC . (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................CTGGGAGAAGAGTGGTGAAGA.......................................................................................... | 21 | 1 | 50.00 | 50.00 | - | - | 2.00 | 7.00 | - | - | - | - | - | 2.00 | - | 1.00 | 1.00 | - | 3.00 | 1.00 | 4.00 | - | - | 1.00 | - | 1.00 | - | - | - | - | - | 3.00 | 1.00 | - | - | - | 1.00 | - | 2.00 | - | 1.00 | - | 2.00 | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 2.00 | 1.00 | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................TTCGCCACTTCCCTCCCTGCAG.................................................. | 22 | 1 | 43.00 | 43.00 | - | - | - | - | 3.00 | 8.00 | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | 2.00 | 1.00 | - | - | - | 1.00 | - | 1.00 | - | - | 3.00 | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | 1.00 | 1.00 | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | 1.00 | 1.00 | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | 1.00 | - | - | - |
| .....................................................................................................................................................TTCGCCACTTCCCTCCCTGCAGT................................................. | 23 | 1 | 28.00 | 43.00 | - | - | - | 1.00 | 5.00 | 3.00 | - | - | - | - | 4.00 | - | - | - | - | - | - | 1.00 | - | - | 2.00 | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - |
| ..............................................................................................................CTGGGAGAAGAGTGGTGAAG........................................................................................... | 20 | 1 | 23.00 | 23.00 | - | 5.00 | - | 3.00 | - | - | - | - | - | - | - | 3.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | 2.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................ACGGCTGGGAGAAGAGTGGTGA............................................................................................. | 22 | 1 | 20.00 | 20.00 | - | 3.00 | 7.00 | - | - | - | - | - | 4.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................TTCGCCACTTCCCTCCCTGCAGA................................................. | 23 | 1 | 14.00 | 14.00 | - | - | - | - | 6.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................TTCGCCACTTCCCTCCCTGC.................................................... | 20 | 1 | 14.00 | 14.00 | - | 1.00 | - | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | 1.00 | 2.00 | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................CTGGGAGAAGAGTGGTGAAGAA......................................................................................... | 22 | 1 | 12.00 | 12.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 2.00 | - | 1.00 | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................CGGCTGGGAGAAGAGTGGTGA............................................................................................. | 21 | 1 | 12.00 | 12.00 | 4.00 | - | - | - | - | - | 2.00 | - | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TGGGAGAAGAGTGGTGAAGA.......................................................................................... | 20 | 2 | 8.50 | 8.50 | - | - | - | 1.00 | - | 1.50 | - | 0.50 | 1.00 | 0.50 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.50 | 0.50 | - |
| ............................................................................................................GGCTGGGAGAAGAGTGGTGA............................................................................................. | 20 | 1 | 7.00 | 7.00 | 1.00 | - | - | - | - | - | 2.00 | 1.00 | - | - | 1.00 | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................CGGCTGGGAGAAGAGTGGT............................................................................................... | 19 | 1 | 6.00 | 6.00 | 4.00 | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................TTCGCCACTTCCCTCCCTG..................................................... | 19 | 1 | 6.00 | 6.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................GGCTGGGAGAAGAGTGGTGAAG........................................................................................... | 22 | 1 | 6.00 | 6.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................GGCTGGGAGAAGAGTGGTGAA............................................................................................ | 21 | 1 | 5.00 | 5.00 | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................CGGCTGGGAGAAGAGTGGTGAA............................................................................................ | 22 | 1 | 5.00 | 5.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................ACGGCTGGGAGAAGAGTGGTGAA............................................................................................ | 23 | 1 | 5.00 | 5.00 | - | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................CTGGGAGAAGAGTGGTGA............................................................................................. | 18 | 1 | 5.00 | 5.00 | 4.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................GGCTGGGAGAAGAGTGGTG.............................................................................................. | 19 | 1 | 5.00 | 5.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................GGCTGGGAGAAGAGTGGTGAAGA.......................................................................................... | 23 | 1 | 5.00 | 5.00 | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................ACGGCTGGGAGAAGAGTGGTGAT............................................................................................ | 23 | 1 | 4.00 | 20.00 | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................TCGCCACTTCCCTCCCTGCAGT................................................. | 22 | 1 | 4.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................TTCGCCACTTCCCTCCCTGCA................................................... | 21 | 1 | 4.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................CTGGGAGAAGAGTGGTGAAGAAA........................................................................................ | 23 | 1 | 4.00 | 12.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................GCTGGGAGAAGAGTGGTGAAG........................................................................................... | 21 | 1 | 3.00 | 3.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................CTGGGAGAAGAGTGGTGAA............................................................................................ | 19 | 1 | 3.00 | 3.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................CGGCTGGGAGAAGAGTGGTG.............................................................................................. | 20 | 1 | 3.00 | 3.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................GCTGGGAGAAGAGTGGTGA............................................................................................. | 19 | 1 | 3.00 | 3.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ............................................................................................................GGCTGGGAGAAGAGTGGTGAAGAA......................................................................................... | 24 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................TTCGCCACTTCCCTCCCTGCAA.................................................. | 22 | 1 | 3.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................TTCGCCACTTCCCTCCCTGCAAA................................................. | 23 | 1 | 3.00 | 4.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................CGGCTGGGAGAAGAGTGGTGAAG........................................................................................... | 23 | 1 | 3.00 | 3.00 | - | 1.00 | - | - | - | - | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................ACGGCTGGGAGAAGAGTGG................................................................................................ | 19 | 1 | 3.00 | 3.00 | 2.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................ACGGCTGGGAGAAGAGTGGTG.............................................................................................. | 21 | 1 | 3.00 | 3.00 | - | 1.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................CGGCTGGGAGAAGAGTGGTGAAGTT......................................................................................... | 25 | 1 | 3.00 | 3.00 | - | - | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................CGGCTGGGAGAAGAGTGGTGAAGA.......................................................................................... | 24 | 1 | 3.00 | 3.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................GCTGGGAGAAGAGTGGTGAAGA.......................................................................................... | 22 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................GCTGGGAGAAGAGTGGTGAAA........................................................................................... | 21 | 1 | 2.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................TTCGCCACTTCCCTCCCTGCAGTT................................................ | 24 | 1 | 2.00 | 43.00 | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................ACGGCTGGGAGAAGAGTGGTGAAT........................................................................................... | 24 | 1 | 2.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - |
| ...........................................................................................................CGGCTGGGAGAAGAGTGGTGAAA........................................................................................... | 23 | 1 | 2.00 | 5.00 | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................GCCACTTCCCTCCCTGCAGA................................................. | 20 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................ACGGCTGGGAGAAGAGTGGT............................................................................................... | 20 | 1 | 2.00 | 2.00 | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................CGGCTGGGAGAAGAGTGG................................................................................................ | 18 | 1 | 2.00 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................TTCGCCACTTCCCTCCCTGCAGAA................................................ | 24 | 1 | 2.00 | 14.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TGGGAGAAGAGTGGTGAAGAA......................................................................................... | 21 | 1 | 2.00 | 2.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TGGGAGAAGAGTGGTGAA............................................................................................ | 18 | 3 | 1.67 | 1.67 | 0.67 | 0.67 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.33 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................CGGACGGCTGGGA......................................................................................................... | 13 | 7 | 1.14 | 1.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | 0.14 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 | 0.14 | 0.14 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.14 |
| ..............................................................................................................CTGGGAGAAGAGTGGTGAAGACT........................................................................................ | 23 | 1 | 1.00 | 50.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................GCTGGGAGAAGAGTGGTGAAGT.......................................................................................... | 22 | 1 | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................GCTGGGAGAAGAGTGGTGAAGAA......................................................................................... | 23 | 1 | 1.00 | 1.00 | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................CTGGGAGAAGAGTGGTGAAGTAA........................................................................................ | 23 | 1 | 1.00 | 23.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................TTCGCCACTTCCCTCCCTGCATT................................................. | 23 | 1 | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TGGGAGAAGAGTGGTGAAGG.......................................................................................... | 20 | 2 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................................................................AGGGTGCCAGCTTCAATGACA........... | 21 | 1 | 1.00 | 1.00 | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...................................................................................................................................AGAGTATTGATTGTGCTG........................................................................ | 18 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................TTCGCCACTTCCCTCCCTGCAGTTA............................................... | 25 | 1 | 1.00 | 43.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................................................................................................................TCGCCACTTCCCTCCCTGCAG.................................................. | 21 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ........................................................................................................................................................GCCACTTCCCTCCCTGCAA.................................................. | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................CTGGGAGAAGAGTGGTGACG........................................................................................... | 20 | 1 | 1.00 | 5.00 | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................GCTGGGAGAAGAGTGGTGAA............................................................................................ | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TGGGAGAAGAGTGGTGAAG........................................................................................... | 19 | 2 | 1.00 | 1.00 | - | - | - | 0.50 | - | - | - | 0.50 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................TTCGCCACTTCCCTCCCTGCCG.................................................. | 22 | 1 | 1.00 | 14.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................ACGGCTGGGAGAAGAGTGGTGAAA........................................................................................... | 24 | 1 | 1.00 | 5.00 | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................CGGCTGGGAGAAGAGTA................................................................................................. | 17 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................................................................................................AAGGGTGCCAGCTTCAATG.............. | 19 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .......................................................................................................................................................CGCCACTTCCCTCCCTGCAGT................................................. | 21 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ............................................................................................................GGCTGGGAGAAGAGTGGTGAG............................................................................................ | 21 | 1 | 1.00 | 7.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................CGGCTGGGAGAAGAGTGGTGAAGT.......................................................................................... | 24 | 1 | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................ACGGCTGGGAGAAGAGTGGTGAAGA.......................................................................................... | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................TTCGCCACTTCCCTCCCTGCAGTTT............................................... | 25 | 1 | 1.00 | 43.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................TTCGCCACTTCCCTCCCTGCTGTC................................................ | 24 | 1 | 1.00 | 14.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - |
| ..............................................................................................................CTGGGAGAAGAGTGGTGAAA........................................................................................... | 20 | 1 | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...........................................................................................................CGGCTGGGAGAAGAGTGGTGATT........................................................................................... | 23 | 1 | 1.00 | 12.00 | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................TTCGCCACTTCCCTCCCTGCT................................................... | 21 | 1 | 1.00 | 14.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................CTGGGAGAAGAGTGGTGAAAG.......................................................................................... | 21 | 1 | 1.00 | 3.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .....................................................................................................................................................TTCGCCACTTCCCTCCCTGCAT.................................................. | 22 | 1 | 1.00 | 4.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..........................................................................................................ACGGCTGGGAGAAGAGTGGTGAG............................................................................................ | 23 | 1 | 1.00 | 20.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ..............................................................................................................CTGGGAGAAGAGTGGTGAGG........................................................................................... | 20 | 1 | 1.00 | 5.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ...............................................................................................................TGGGAGAAGAGTGGTGA............................................................................................. | 17 | 5 | 0.20 | 0.20 | - | 0.20 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| .............................................................................................................................................TTGTGCTGTTCGC................................................................... | 13 | 10 | 0.10 | 0.10 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 0.10 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| CACCTTCCAGTATTTCATCTGCATTGGCATCCCACCTGCTCCTTGCCGAGGTGAGCCTGCCCTTCCCTTGAGCCGTCTACTTTTAGAAGAAGCCATTACTGAGCGGACGGCTGGGAGAAGAGTGGTGAAGAAGAGTATTGATTGTGCTGTTCGCCACTTCCCTCCCTGCAGATTACCCGTGGAGATTCAAGGGTGCCAGCTTCAATGACAACATCATAAAG .............................................................................................................((.(((((..(((((((...(((.(((((.....))))).))))))))))..))))).)).................................................... .........................................................................................................106..............................................................171................................................ | Size | Perfect hit | Total Norm | Perfect Norm | SRR207111(GSM721073) Whole cell RNA. (cell line) | SRR207115(GSM721077) XRN1&2 knockdown. (XRN1/XRN2 cell line) | SRR039615(GSM531978) Severe Chronic Hepatitis B Liver Tissue. (liver) | SRR039625(GSM531988) HBV(-) HCV(-) HCC Tissue Sample. (liver) | SRR207114(GSM721076) IP against AGO 1 & 2, RRP40 knockdown. (ago1/2 RRP40 cell line) | SRR387910(GSM843862) antibody for ip: Ago2. (ago2 cell line) | SRR033716(GSM497061) Mentle Cell Lymphoma (MCL114). (B cell) | SRR207112(GSM721074) RRP40 knockdown. (RRP40 cell line) | SRR039614(GSM531977) HBV-infected Liver Tissue. (liver) | SRR107296(GSM677703) 18-30nt fraction of small RNA. (cell line) | SRR029124(GSM416753) HeLa. (hela) | SRR033728(GSM497073) MALT (MALT413). (B cell) | SRR039613(GSM531976) Human Normal Liver Tissue Sample 3. (liver) | SRR033718(GSM497063) Multiple Myeloma (U266). (B cell) | SRR039612(GSM531975) Human Normal Liver Tissue Sample 2. (liver) | SRR039611(GSM531974) Human Normal Liver Tissue Sample 1. (liver) | SRR033729(GSM497074) splenic MZL (Splenic414). (B cell) | SRR330921(SRX091759) tissue: normal skindisease state: normal. (skin) | SRR207113(GSM721075) IP against AGO 1 & 2. (ago1/2 cell line) | SRR330896(SRX091734) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330901(SRX091739) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330922(SRX091760) tissue: normal skindisease state: normal. (skin) | SRR330894(SRX091732) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR040035(GSM532920) G001T. (cervix) | SRR330895(SRX091733) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330893(SRX091731) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330865(SRX091703) tissue: skin psoriatic involveddisease state:. (skin) | SRR033727(GSM497072) HIV-positive DL (HIV412). (B cell) | SRR330890(SRX091728) tissue: skin psoriatic uninvolveddisease stat. (skin) | GSM450608(GSM450608) miRNA sequencing raw reads from post-mortem s. (brain) | SRR207116(GSM721078) Nuclear RNA. (cell line) | SRR037944(GSM510482) 293DcrTN_cand5. (cell line) | SRR040014(GSM532899) G623N. (cervix) | SRR039616(GSM531979) HBV(+) Distal Tissue Sample 1. (liver) | SRR033712(GSM497057) Burkitt Lymphoma (BL510). (B cell) | SRR330867(SRX091705) tissue: skin psoriatic involveddisease state:. (skin) | SRR330889(SRX091727) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR444057(SRX128905) Sample 15cDNABarcode: AF-PP-334: ACG CTC TTC . (skin) | SRR039617(GSM531980) HBV(+) Adjacent Tissue Sample 1. (liver) | SRR330884(SRX091722) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330902(SRX091740) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330903(SRX091741) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330906(SRX091744) tissue: normal skindisease state: normal. (skin) | SRR330904(SRX091742) tissue: normal skindisease state: normal. (skin) | SRR207118(GSM721080) RRP40 knockdown. (RRP40 cell line) | TAX577579(Rovira) total RNA. (breast) | SRR330900(SRX091738) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR553572(SRX182778) source: Frontal Cortex. (Frontal Cortex) | SRR038861(GSM458544) MM466. (cell line) | TAX577739(Rovira) total RNA. (breast) | GSM450606(GSM450606) miRNA sequencing raw reads from post-mortem s. (brain) | SRR363675(GSM830252) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR330913(SRX091751) tissue: normal skindisease state: normal. (skin) | SRR330917(SRX091755) tissue: normal skindisease state: normal. (skin) | SRR330915(SRX091753) tissue: normal skindisease state: normal. (skin) | SRR330898(SRX091736) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR039618(GSM531981) HBV(+) Side Tissue Sample 1. (liver) | SRR330891(SRX091729) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR038856(GSM458539) D11. (cell line) | SRR330923(SRX091761) tissue: normal skindisease state: normal. (skin) | SRR343333(GSM796036) KSHV (HHV8). (cell line) | TAX577744(Rovira) total RNA. (breast) | SRR039624(GSM531987) HBV(-) HCV(-) Adjacent Tissue Sample. (liver) | TAX577746(Rovira) total RNA. (breast) | TAX577589(Rovira) total RNA. (breast) | SRR189782 | SRR038855(GSM458538) D10. (cell line) | SRR330912(SRX091750) tissue: normal skindisease state: normal. (skin) | SRR330908(SRX091746) tissue: normal skindisease state: normal. (skin) | SRR330919(SRX091757) tissue: normal skindisease state: normal. (skin) | SRR330880(SRX091718) tissue: skin psoriatic involveddisease state:. (skin) | SRR330875(SRX091713) tissue: skin psoriatic involveddisease state:. (skin) | TAX577741(Rovira) total RNA. (breast) | SRR330874(SRX091712) tissue: skin psoriatic involveddisease state:. (skin) | SRR444053(SRX128901) Sample 13cDNABarcode: AF-PP-341: ACG CTC TTC . (skin) | GSM956925AGO2Paz8(GSM956925) cell line: HEK293 cell linepassages: 15-20ip . (ago2 cell line) | SRR444059(SRX128907) Sample 17cDNABarcode: AF-PP-339: ACG CTC TTC . (skin) | SRR330916(SRX091754) tissue: normal skindisease state: normal. (skin) | SRR033730(GSM497075) Lymphoblastic (ALL411). (B cell) | SRR094132(GSM651908) small RNA(18-35nt)small RNA deep sequencing o. (uterus) | SRR191549(GSM715659) 105genomic small RNA (size selected RNA from . (breast) | GSM450599(GSM450599) miRNA sequencing raw reads from post-mortem s. (brain) | SRR363673(GSM830250) cell type: human foreskin fibroblasts (HFF)tr. (fibroblast) | SRR191500(GSM715610) 7genomic small RNA (size selected RNA from to. (breast) | TAX577588(Rovira) total RNA. (breast) | SRR330870(SRX091708) tissue: skin psoriatic involveddisease state:. (skin) | SRR191418(GSM715528) 40genomic small RNA (size selected RNA from t. (breast) | SRR330897(SRX091735) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191419(GSM715529) 41genomic small RNA (size selected RNA from t. (breast) | SRR038863(GSM458546) MM603. (cell line) | SRR342896(SRX096792) small RNA seq of Right atrial tissue. (heart) | GSM450605(GSM450605) miRNA sequencing raw reads from post-mortem s. (brain) | TAX577742(Rovira) total RNA. (breast) | SRR343336 | SRR040010(GSM532895) G529N. (cervix) | SRR330914(SRX091752) tissue: normal skindisease state: normal. (skin) | SRR330878(SRX091716) tissue: skin psoriatic involveddisease state:. (skin) | DRR000561(DRX000319) Isolation of RNA following immunoprecipitatio. (ago2 cell line) | SRR039620(GSM531983) HBV(+) Adjacent Tissue Sample 2. (liver) | GSM956925Ago2PAZ(GSM956925) cell line: HEK293 cell linepassages: 15-20ip . (ago2 cell line) | SRR038854(GSM458537) MM653. (cell line) | SRR191551(GSM715661) 32genomic small RNA (size selected RNA from t. (breast) | SRR191602(GSM715712) 60genomic small RNA (size selected RNA from t. (breast) | SRR060981(GSM569185) Human centroblast [09-001]. (cell line) | SRR343337 | GSM359198(GSM359198) hepg2_cip_tap_pasr_a. (cell line) | SRR330907(SRX091745) tissue: normal skindisease state: normal. (skin) | SRR330892(SRX091730) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR330866(SRX091704) tissue: skin psoriatic involveddisease state:. (skin) | SRR191625(GSM715735) 32genomic small RNA (size selected RNA from t. (breast) | SRR039637(GSM518474) THP1_total_sRNAs. (cell line) | SRR040017(GSM532902) G645T. (cervix) | SRR040036(GSM532921) G243N. (cervix) | SRR189780(GSM714640) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR191578(GSM715688) 100genomic small RNA (size selected RNA from . (breast) | SRR330868(SRX091706) tissue: skin psoriatic involveddisease state:. (skin) | SRR330918(SRX091756) tissue: normal skindisease state: normal. (skin) | SRR330863(SRX091701) tissue: skin psoriatic involveddisease state:. (skin) | GSM359203(GSM359203) hepg2_untreated_b. (cell line) | SRR330877(SRX091715) tissue: skin psoriatic involveddisease state:. (skin) | SRR191605(GSM715715) 76genomic small RNA (size selected RNA from t. (breast) | SRR038862(GSM458545) MM472. (cell line) | SRR330911(SRX091749) tissue: normal skindisease state: normal. (skin) | SRR191554(GSM715664) 99genomic small RNA (size selected RNA from t. (breast) | SRR039621(GSM531984) HBV(+) HCC Tissue Sample 2. (liver) | SRR330881(SRX091719) tissue: skin psoriatic uninvolveddisease stat. (skin) | SRR191580(GSM715690) 103genomic small RNA (size selected RNA from . (breast) | GSM450603(GSM450603) miRNA sequencing raw reads from post-mortem s. (brain) | SRR189777(GSM714637) cell line: HEK293clip variant: PAR-CLIPenzyma. (cell line) | SRR191603(GSM715713) 71genomic small RNA (size selected RNA from t. (breast) | SRR330861(SRX091699) tissue: skin psoriatic involveddisease state:. (skin) | DRR000556(DRX000314) THP-1 whole cell RNA, after 3 day treatment w. (cell line) | SRR330860(SRX091698) tissue: skin psoriatic involveddisease state:. (skin) | SRR040015(GSM532900) G623T. (cervix) | SRR040027(GSM532912) G220T. (cervix) | SRR444052(SRX128900) Sample 12cDNABarcode: AF-PP-340: ACG CTC TTC . (skin) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................GGTGAAGAAGAGTATTTT................................................................................ | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | |
| .......................................................................................................................................................CGCCACTTCCCTCCCGAG.................................................... | 18 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ....................................................GAGCCTGCCCTTCCCGACC...................................................................................................................................................... | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| ..............................................................................................................................................TGTGCTGTTCGCCACTGGC............................................................ | 19 | 1.00 | 0.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
| .......................................................................................................................GAGTGGTGAAGAAGAGTATTGATTG............................................................................. | 25 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| ......................................................GCCTGCCCTTCCCTTGAGCC................................................................................................................................................... | 20 | 1 | 1.00 | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 1.00 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - |