| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3R:12681982-12682079 + | CGATTCTGTGCGACCTCGTAAACGTATACTGAATGTATCCTGAGTGTATCCTATCCGGTATACCTTCAGTATACGTAACACGAGAGAGCACTAACAAC |
| droSim1 | chr3R:8791202-8791299 - | CGATTCTGTGCGACCTCGTAAACGTATACTGAATGTATCCTGAGTGTATCCTATCCGGTATACCTTCAGTATACGTAACACGAGATAGCACGAGCAAC |
| droSec1 | super_12:1472230-1472327 - | CGATTCTGTGCGACCTCGTAAACGTATACTGAATGTATCCTGAGTGTATCCTATCCGGTATACCTTCAGTATACGTAACACGAGATAGCACGAGCAAC |
| droYak2 | chr3R:11708953-11709050 - | CGATTCTGTGCGACCTCGTAAACGTATACTGAATGTATCCTGAGTGTATCCTATCCGGTATACCTTCAGTATACGTAACACGAGAAAGCACGAGCAAC |
| droEre2 | scaffold_4770:8952402-8952499 - | CGATTCTGTGCGACCTCGTAAACGTATACTGAATGTATCCTGAGTGTATCCTATCCGGTATACCTTCAGTATACGTAACACGAGAAAGCACGAGCAAC |
| droAna3 | scaffold_13340:7215188-7215285 - | TGATTTTGTGCAACTTCGTATACGTATACTGAATGTATCCTGAGTGTATCCTATCCGGTATACCTTCAGTATACGTAACACGAGGTAACACCTTCAAC |
| dp4 | chr2:17671752-17671849 + | -GAACCTGTGCGACTTCGTATACGTATACTGAATGTATCCTGAGTGTATCCTATCCGGTATACCTTCAGTATACGTAACACGAGGAAACACCAGCAAC |
| droPer1 | super_0:11526558-11526655 + | -GAACCTGTGCGACTTCGTATACGTATACTGAATGTATCCTGAGTGTATCCTATCCGGTATACCTTCAGTATACGTAACACGAGGAAACACCAGCAAC |
| droWil1 | scaffold_181130:4905585-4905682 - | TGAATCTGTGCAACTTCGTATACGTATACTGAATGTATCCTGAGTGTATGCTATCCGGTATACCTTCAGTATACGTAACACGATGAAACACCAACAAC |
| droVir3 | scaffold_12855:414691-414788 - | AGAGCCTGTGCAACTTCGTATACGTATACTGAATGTATCCTGAGTGTATCCTATCCGGTATACCTTCAGTATACGTAACACGAGGAAACACCAGCGAC |
| droMoj3 | scaffold_6540:1943729-1943826 + | TGAGCCTGTGCAACTTCGTATACGTATACTGAATGTATCCTGAGTGTATCCTATCCGGTATACCTTCAGTATACGTAACACGAGGAAACACCAGCAAC |
| droGri2 | scaffold_14906:12815108-12815201 + | -----CTGTGCAACTTCGTATACGTATACTGAATGTATCCTGAGTGTATCCTATCCGGTATACCTTCAGTATACGTAACACGAGGAAACACCAGCAAC |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3R:12681982-12682079 + | CGATTCTGTGCGACCTCGTAAACGTATACTGAATGTATCCTGAGTGTATCCTATCCGGTATACCTTCAGTATACGTAACACGAGAGAGCACTAACAAC |
| droSim1 | chr3R:8791202-8791299 - | CGATTCTGTGCGACCTCGTAAACGTATACTGAATGTATCCTGAGTGTATCCTATCCGGTATACCTTCAGTATACGTAACACGAGATAGCACGAGCAAC |
| droSec1 | super_12:1472230-1472327 - | CGATTCTGTGCGACCTCGTAAACGTATACTGAATGTATCCTGAGTGTATCCTATCCGGTATACCTTCAGTATACGTAACACGAGATAGCACGAGCAAC |
| droYak2 | chr3R:11708953-11709050 - | CGATTCTGTGCGACCTCGTAAACGTATACTGAATGTATCCTGAGTGTATCCTATCCGGTATACCTTCAGTATACGTAACACGAGAAAGCACGAGCAAC |
| droEre2 | scaffold_4770:8952402-8952499 - | CGATTCTGTGCGACCTCGTAAACGTATACTGAATGTATCCTGAGTGTATCCTATCCGGTATACCTTCAGTATACGTAACACGAGAAAGCACGAGCAAC |
| droAna3 | scaffold_13340:7215188-7215285 - | TGATTTTGTGCAACTTCGTATACGTATACTGAATGTATCCTGAGTGTATCCTATCCGGTATACCTTCAGTATACGTAACACGAGGTAACACCTTCAAC |
| dp4 | chr2:17671752-17671849 + | -GAACCTGTGCGACTTCGTATACGTATACTGAATGTATCCTGAGTGTATCCTATCCGGTATACCTTCAGTATACGTAACACGAGGAAACACCAGCAAC |
| droPer1 | super_0:11526558-11526655 + | -GAACCTGTGCGACTTCGTATACGTATACTGAATGTATCCTGAGTGTATCCTATCCGGTATACCTTCAGTATACGTAACACGAGGAAACACCAGCAAC |
| droWil1 | scaffold_181130:4905585-4905682 - | TGAATCTGTGCAACTTCGTATACGTATACTGAATGTATCCTGAGTGTATGCTATCCGGTATACCTTCAGTATACGTAACACGATGAAACACCAACAAC |
| droVir3 | scaffold_12855:414691-414788 - | AGAGCCTGTGCAACTTCGTATACGTATACTGAATGTATCCTGAGTGTATCCTATCCGGTATACCTTCAGTATACGTAACACGAGGAAACACCAGCGAC |
| droMoj3 | scaffold_6540:1943729-1943826 + | TGAGCCTGTGCAACTTCGTATACGTATACTGAATGTATCCTGAGTGTATCCTATCCGGTATACCTTCAGTATACGTAACACGAGGAAACACCAGCAAC |
| droGri2 | scaffold_14906:12815108-12815201 + | -----CTGTGCAACTTCGTATACGTATACTGAATGTATCCTGAGTGTATCCTATCCGGTATACCTTCAGTATACGTAACACGAGGAAACACCAGCAAC |
| Species | Read pileup | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
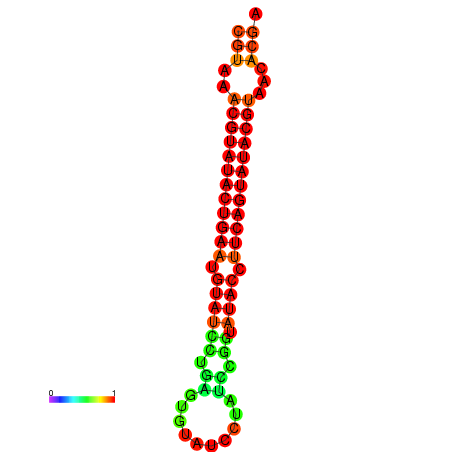
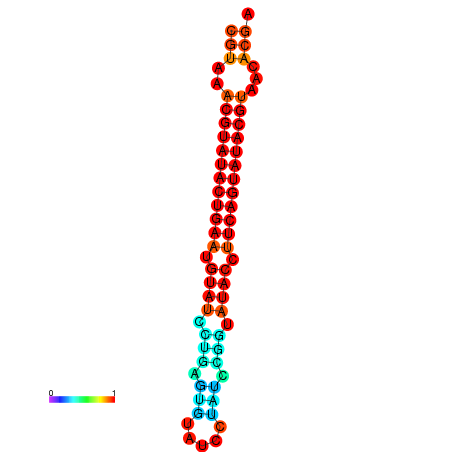
| dG=-24.4, p-value=0.009901 | dG=-24.0, p-value=0.009901 |
|---|---|
 |
 |
| dG=-24.4, p-value=0.009901 | dG=-24.0, p-value=0.009901 |
|---|---|
 |
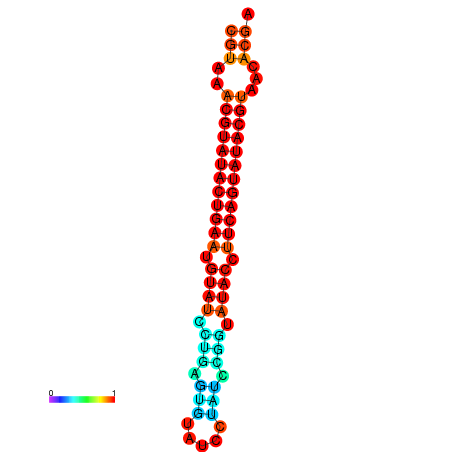 |
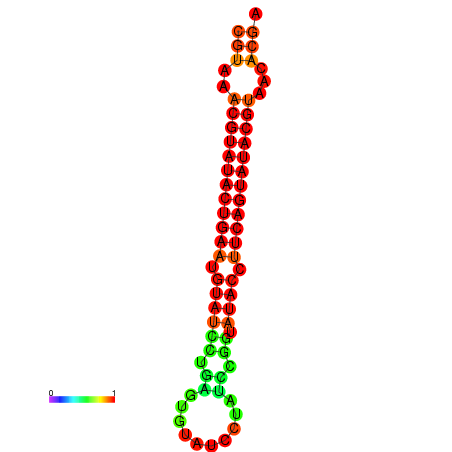
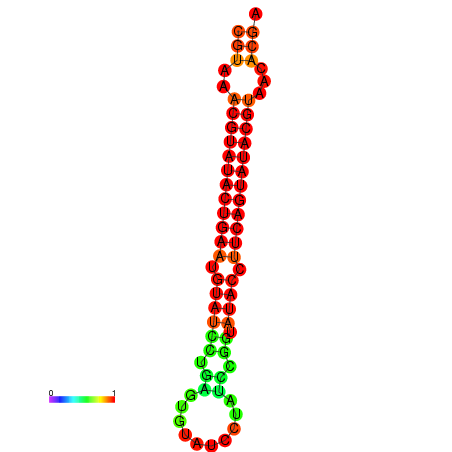
| dG=-24.4, p-value=0.009901 | dG=-24.0, p-value=0.009901 |
|---|---|
 |
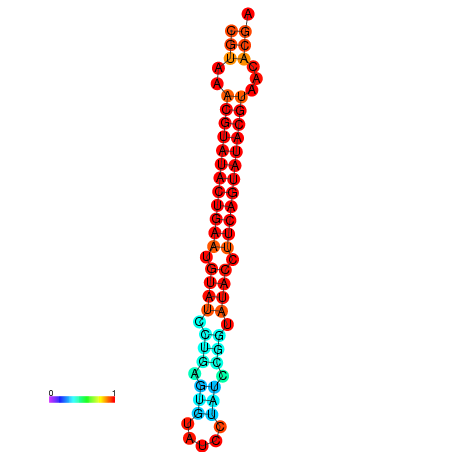 |
| dG=-24.4, p-value=0.009901 | dG=-24.0, p-value=0.009901 |
|---|---|
 |
 |
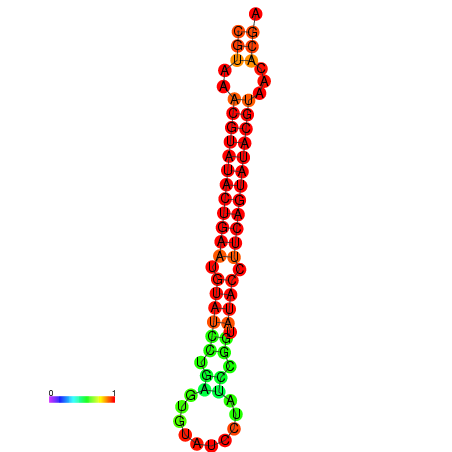
| dG=-24.4, p-value=0.009901 | dG=-24.0, p-value=0.009901 |
|---|---|
 |
 |
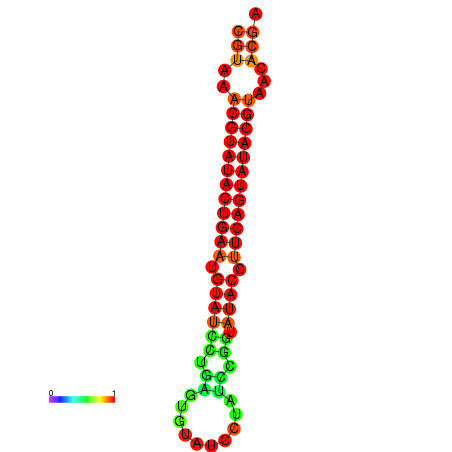
| dG=-26.0, p-value=0.009901 | dG=-25.6, p-value=0.009901 |
|---|---|
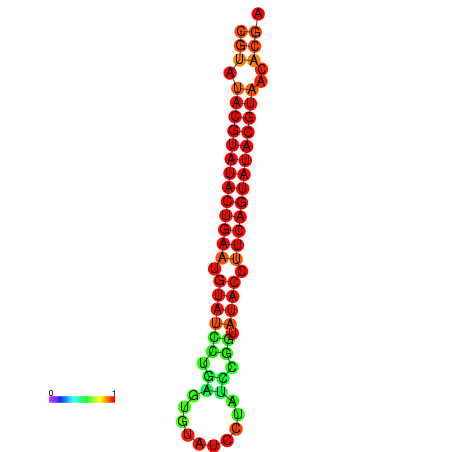 |
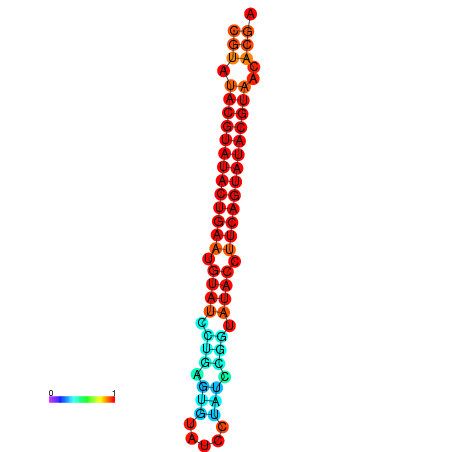 |
| dG=-26.0, p-value=0.009901 | dG=-25.6, p-value=0.009901 |
|---|---|
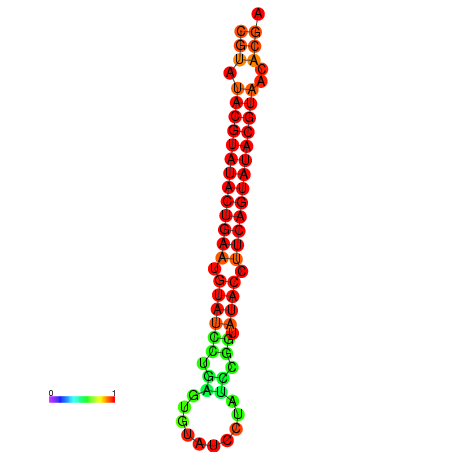 |
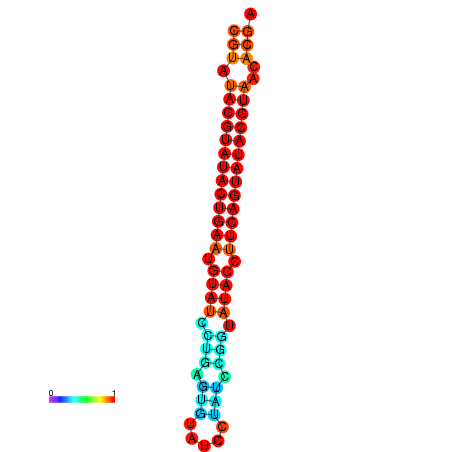 |
| dG=-26.0, p-value=0.009901 | dG=-25.6, p-value=0.009901 |
|---|---|
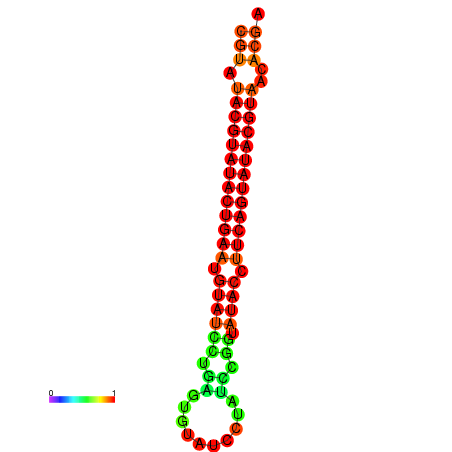 |
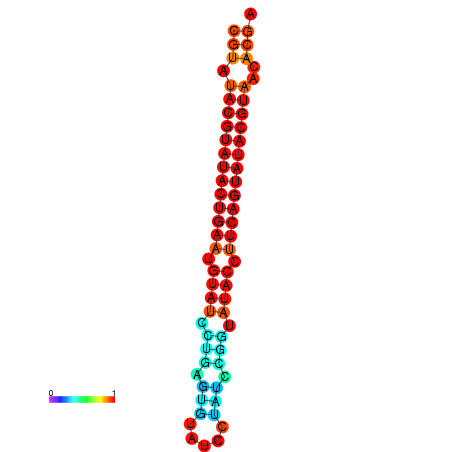 |
| dG=-26.0, p-value=0.009901 | dG=-25.6, p-value=0.009901 | dG=-25.6, p-value=0.009901 | dG=-25.4, p-value=0.009901 | dG=-25.2, p-value=0.009901 |
|---|---|---|---|---|
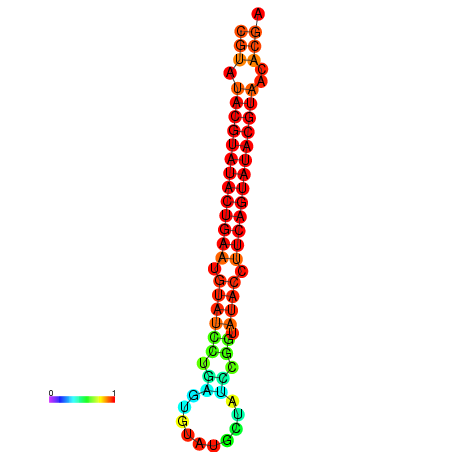 |
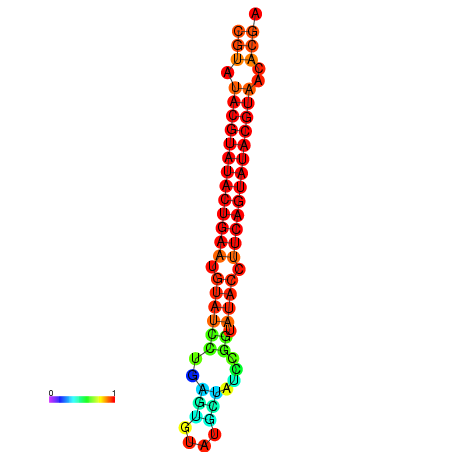 |
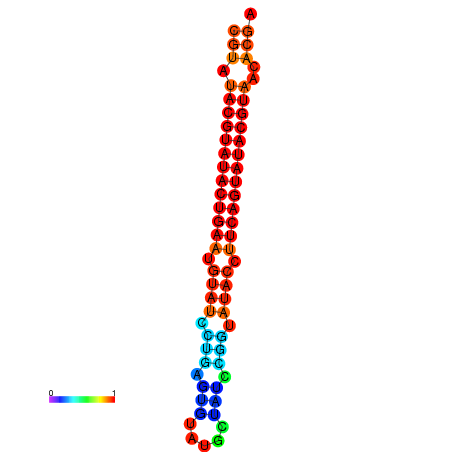 |
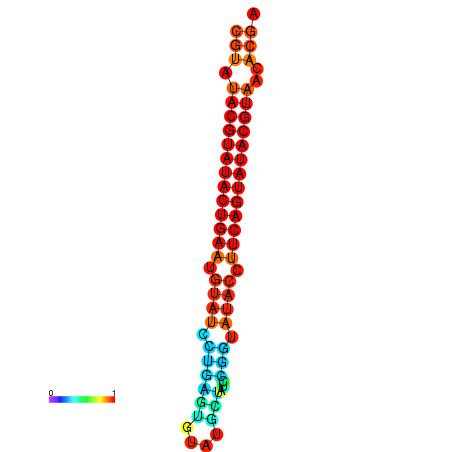 |
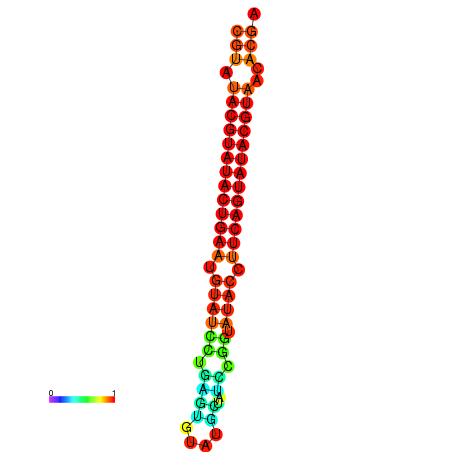 |
| dG=-26.0, p-value=0.009901 | dG=-25.6, p-value=0.009901 |
|---|---|
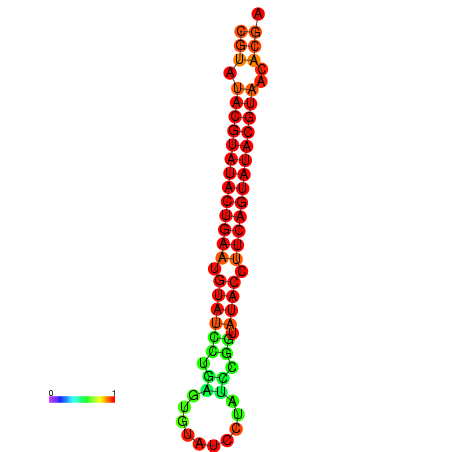 |
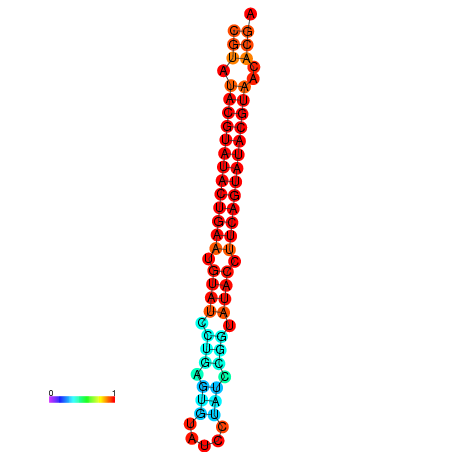 |
| dG=-26.0, p-value=0.009901 | dG=-25.6, p-value=0.009901 |
|---|---|
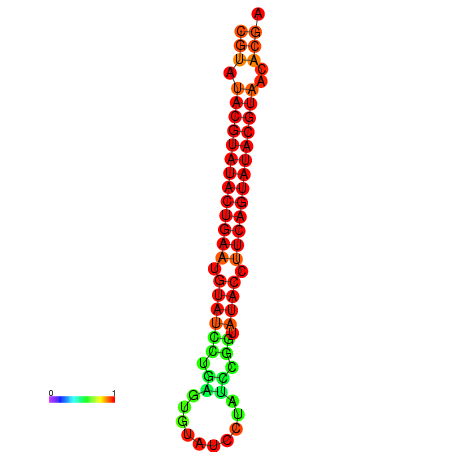 |
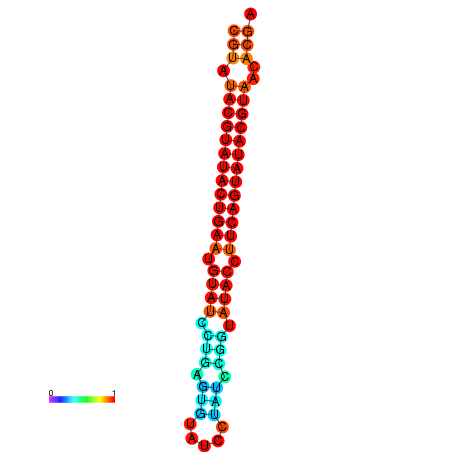 |
| dG=-26.0, p-value=0.009901 | dG=-25.6, p-value=0.009901 |
|---|---|
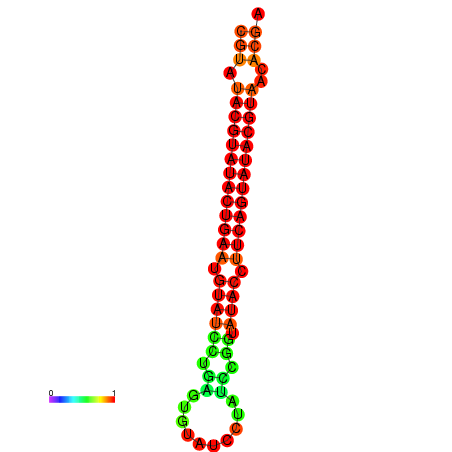 |
 |
Generated: 03/07/2013 at 07:30 PM