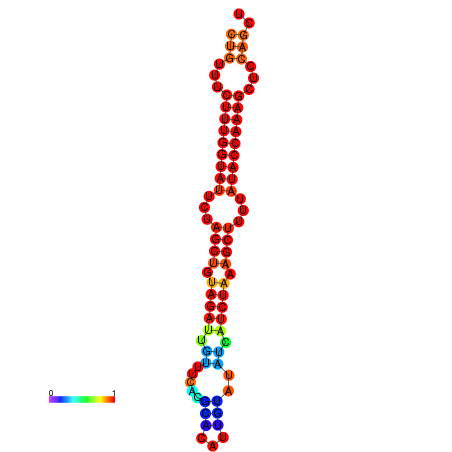
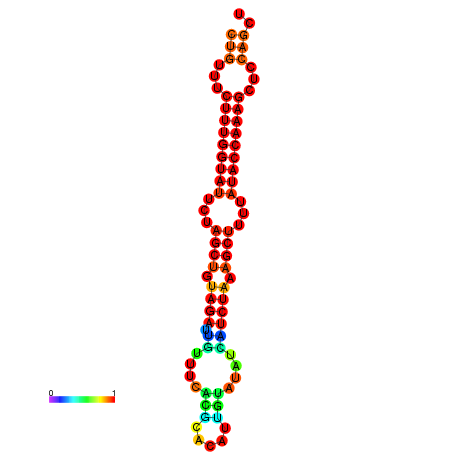
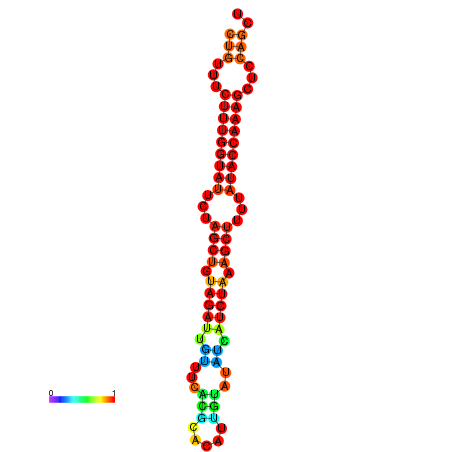
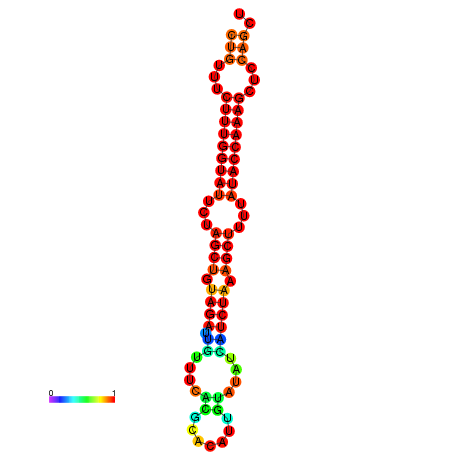
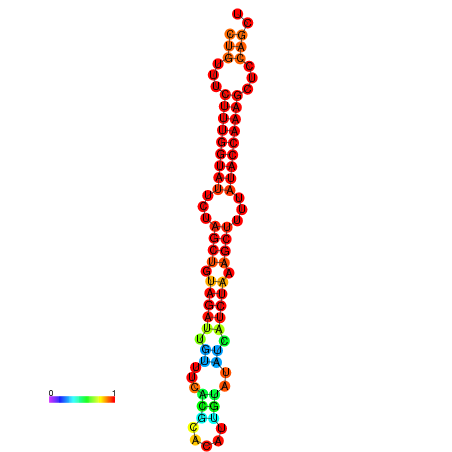
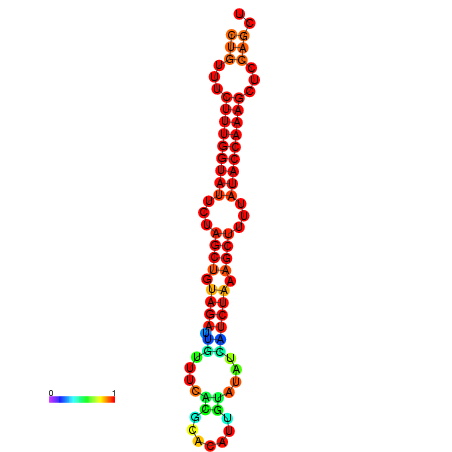
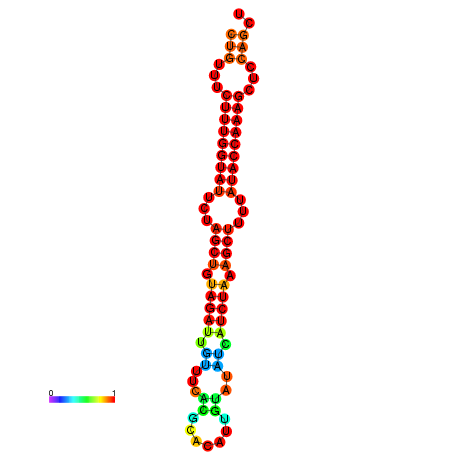
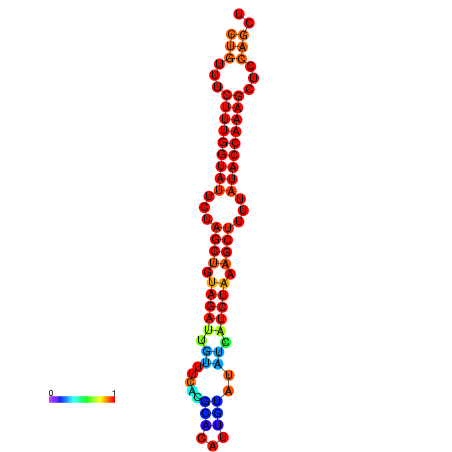
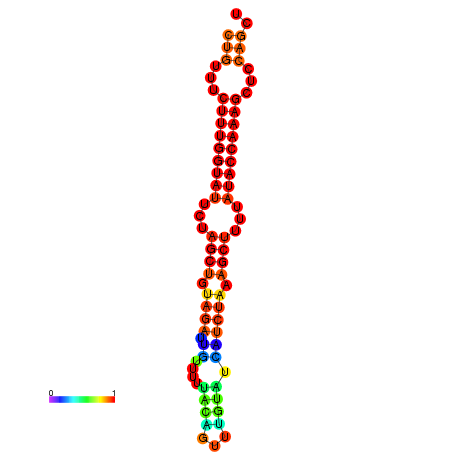
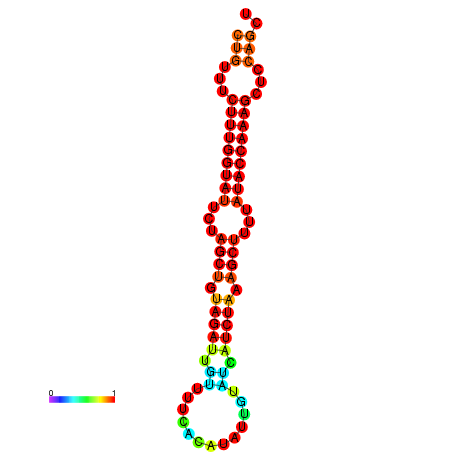
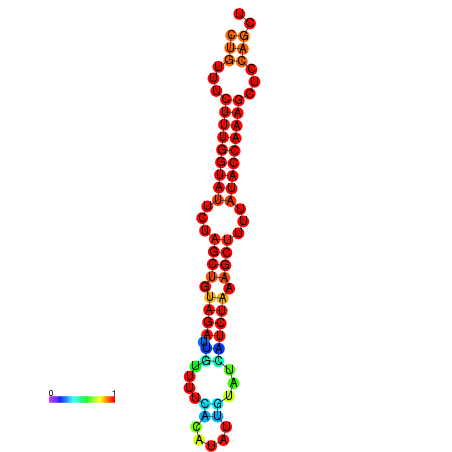
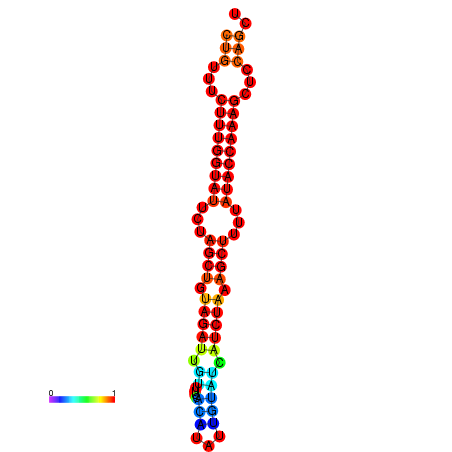
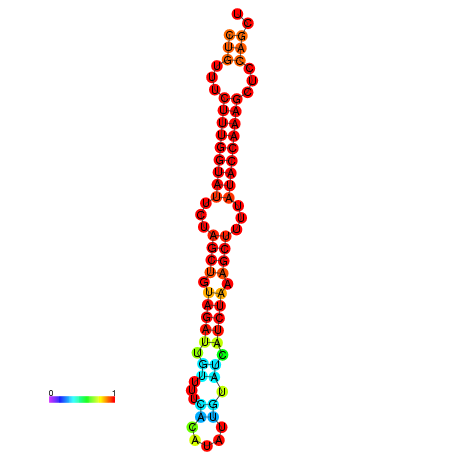
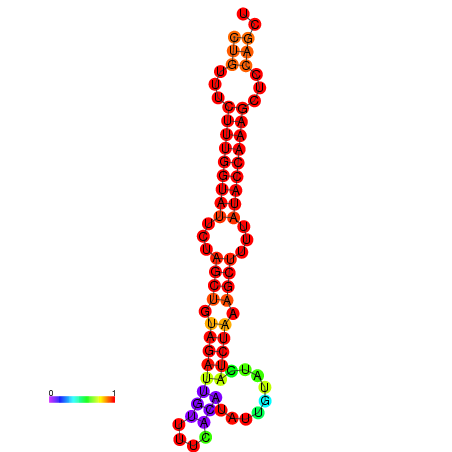
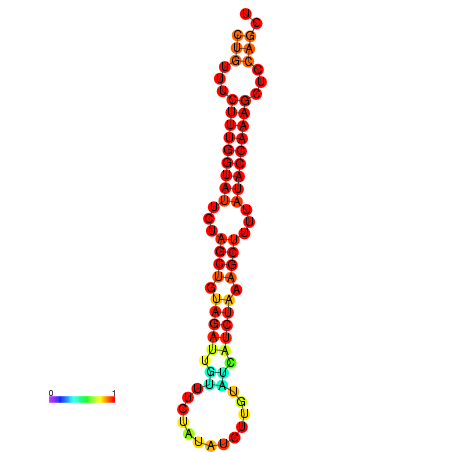
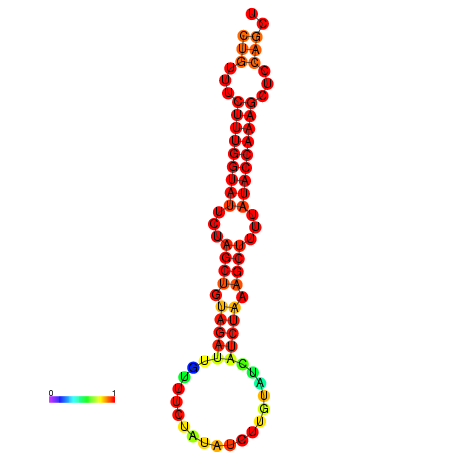| CTCTCCCATTCGCTGTCTGTCGACTTCCATGCACTTTTTGTTCTCCTTTAATTTTTGCTGTTTCTTTGGTATTCTAGCTGTAGATTGTTTTTTGCACATTGTATATCATCTAAAGCTTTTATACCAAAGCTCCAGCTTAAATTGCTTAACA------ | Size | Hit Count | Total Norm | Total | GSM343916
Embryo | GSM444067
Head |
| ..............................................................TCTTTGGTATTCTAGCTGTAG.......................................................................... | 21 | 1 | 7678.00 | 7678 | 6709 | 969 |
| ..............................................................TCTTTGGTATTCTAGCTGTAGA......................................................................... | 22 | 1 | 3324.00 | 3324 | 2969 | 355 |
| ..............................................................TCTTTGGTATTCTAGCTGTA........................................................................... | 20 | 1 | 76.00 | 76 | 47 | 29 |
| ..............................................................TCTTTGGTATTCTAGCTGTAGAT........................................................................ | 23 | 1 | 71.00 | 71 | 63 | 8 |
| ..............................................................TCTTTGGTATTCTAGCTGT............................................................................ | 19 | 1 | 26.00 | 26 | 8 | 18 |
| ..............................................................................................................TAAAGCTTTTATACCAAAGCT.......................... | 21 | 1 | 16.00 | 16 | 11 | 5 |
| ................................................................TTTGGTATTCTAGCTGTAGA......................................................................... | 20 | 1 | 12.00 | 12 | 12 | 0 |
| ................................................................TTTGGTATTCTAGCTGTAG.......................................................................... | 19 | 1 | 10.00 | 10 | 9 | 1 |
| ................................................................TTTGGTATTCTAGCTGTAGATT....................................................................... | 22 | 1 | 8.00 | 8 | 6 | 2 |
| ..............................................................TCTTTGGTATTCTAGCTGTAGATT....................................................................... | 24 | 1 | 7.00 | 7 | 5 | 2 |
| ...............................................................CTTTGGTATTCTAGCTGTAG.......................................................................... | 20 | 1 | 7.00 | 7 | 6 | 1 |
| ...............................................................CTTTGGTATTCTAGCTGTAGA......................................................................... | 21 | 1 | 6.00 | 6 | 6 | 0 |
| ....................................................................................TTGTTTTTTGCACATTGTATATCATC............................................... | 26 | 1 | 5.00 | 5 | 5 | 0 |
| ..............................................................................................................TAAAGCTTTTATACCAAAGCTC......................... | 22 | 1 | 4.00 | 4 | 4 | 0 |
| ...............................................................CTTTGGTATTCTAGCTGTAGAT........................................................................ | 22 | 1 | 4.00 | 4 | 4 | 0 |
| ................................................................TTTGGTATTCTAGCTGTAGAT........................................................................ | 21 | 1 | 4.00 | 4 | 4 | 0 |
| .................................................................TTGGTATTCTAGCTGTAGA......................................................................... | 19 | 1 | 3.00 | 3 | 3 | 0 |
| ...........................................TCCTTTAATTTTTGCTGTT............................................................................................... | 19 | 1 | 2.00 | 2 | 2 | 0 |
| .................................................................TTGGTATTCTAGCTGTAG.......................................................................... | 18 | 1 | 2.00 | 2 | 2 | 0 |
| ..............................................................TCTTTGGTATTCTAGCTG............................................................................. | 18 | 1 | 2.00 | 2 | 1 | 1 |
| .............................................................................................................CTAAAGCTTTTATACCAAAGCT.......................... | 22 | 1 | 2.00 | 2 | 2 | 0 |
| ....................................................................................TTGTTTTTTGCACATTGTATATC.................................................. | 23 | 1 | 1.00 | 1 | 0 | 1 |
| ..............................................................................................................TAAAGCTTTTATACCAAAG............................ | 19 | 1 | 1.00 | 1 | 1 | 0 |
| ............................................................................................................TCTAAAGCTTTTATACCAAAGC........................... | 22 | 1 | 1.00 | 1 | 1 | 0 |