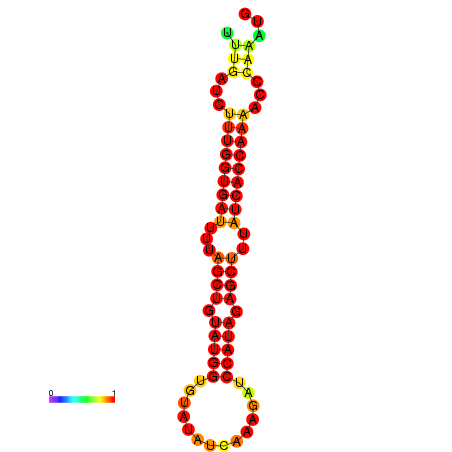
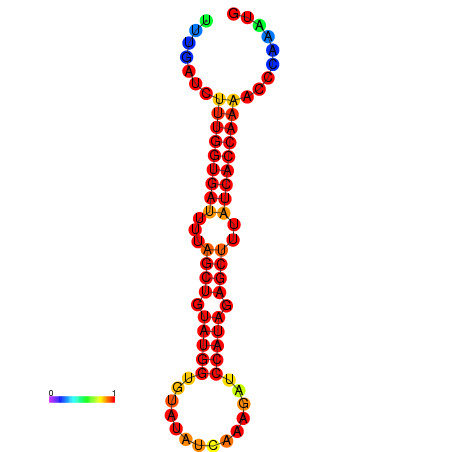
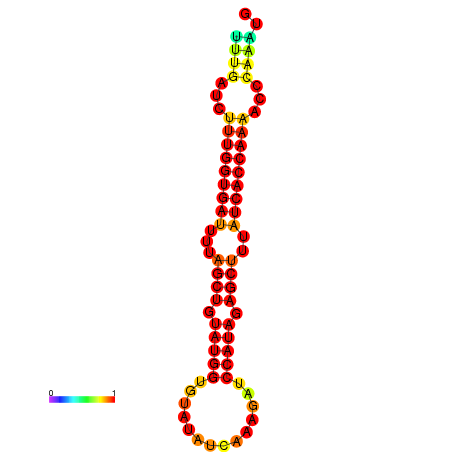
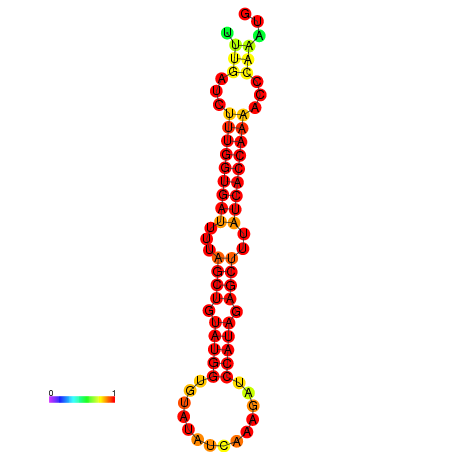
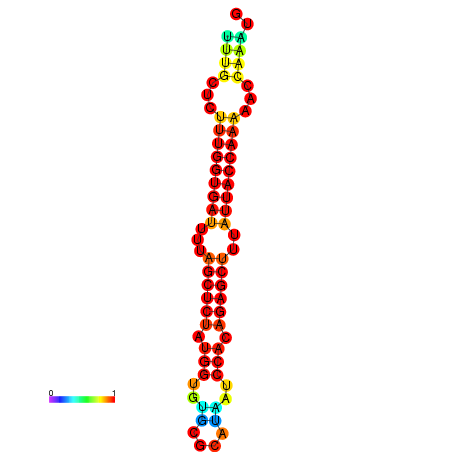
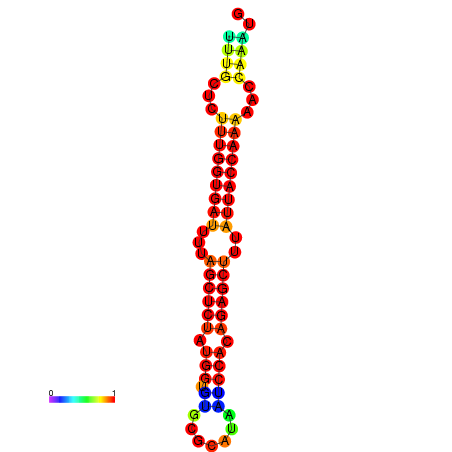
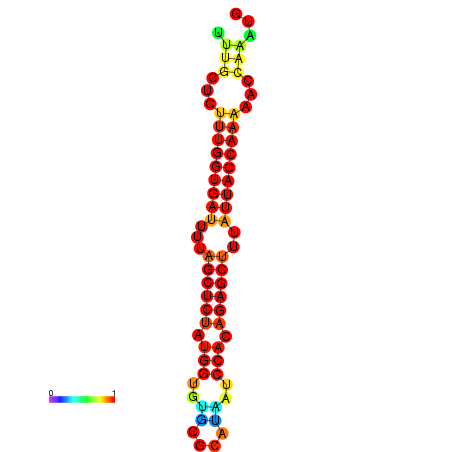
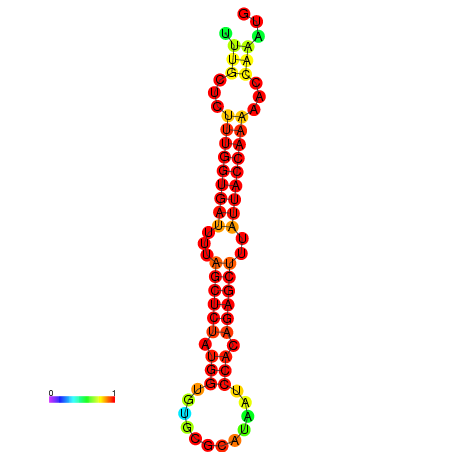
| AGTTGATCTTTTGTTTGCACATTATTTGATCTTTGGTGATTTTAGCTGTATGGTGTATATCAAAGATCCATAGAGCTTTATCACCAAAACCCAAATGGTTTTTGCATTTTTTCGCTTGAGTTGC | Size | Hit Count | Total Norm | Total | GSM343916
Embryo | GSM444067
Head |
| .............................TCTTTGGTGATTTTAGCTGTATG........................................................................ | 23 | 1 | 10038.00 | 10038 | 8752 | 1286 |
| .............................TCTTTGGTGATTTTAGCTGTAT......................................................................... | 22 | 1 | 4862.00 | 4862 | 3232 | 1630 |
| .............................TCTTTGGTGATTTTAGCTGTA.......................................................................... | 21 | 1 | 280.00 | 280 | 161 | 119 |
| .............................TCTTTGGTGATTTTAGCTGTATGG....................................................................... | 24 | 1 | 165.00 | 165 | 153 | 12 |
| ......................................................................TAGAGCTTTATCACCAAAACC................................. | 21 | 1 | 130.00 | 130 | 93 | 37 |
| ......................................................................TAGAGCTTTATCACCAAAACCC................................ | 22 | 1 | 106.00 | 106 | 53 | 53 |
| .............................TCTTTGGTGATTTTAGCTGT........................................................................... | 20 | 1 | 104.00 | 104 | 45 | 59 |
| ..............................CTTTGGTGATTTTAGCTGTATGG....................................................................... | 23 | 1 | 93.00 | 93 | 87 | 6 |
| ..............................CTTTGGTGATTTTAGCTGTATG........................................................................ | 22 | 1 | 44.00 | 44 | 44 | 0 |
| ...............................TTTGGTGATTTTAGCTGTATG........................................................................ | 21 | 1 | 24.00 | 24 | 21 | 3 |
| .............................TCTTTGGTGATTTTAGCTG............................................................................ | 19 | 1 | 15.00 | 15 | 5 | 10 |
| ...............................TTTGGTGATTTTAGCTGTAT......................................................................... | 20 | 1 | 10.00 | 10 | 9 | 1 |
| ......................................................................TAGAGCTTTATCACCAAAAC.................................. | 20 | 1 | 10.00 | 10 | 8 | 2 |
| ................................TTGGTGATTTTAGCTGTATG........................................................................ | 20 | 1 | 5.00 | 5 | 5 | 0 |
| .............................TCTTTGGTGATTTTAGCTGTATGGT...................................................................... | 25 | 1 | 4.00 | 4 | 3 | 1 |
| ..............................CTTTGGTGATTTTAGCTGTAT......................................................................... | 21 | 1 | 4.00 | 4 | 3 | 1 |
| ................................TTGGTGATTTTAGCTGTAT......................................................................... | 19 | 1 | 3.00 | 3 | 3 | 0 |
| .....................................................................ATAGAGCTTTATCACCAAAACC................................. | 22 | 1 | 3.00 | 3 | 3 | 0 |
| ...............................TTTGGTGATTTTAGCTGTATGG....................................................................... | 22 | 1 | 3.00 | 3 | 3 | 0 |
| ......................................................................TAGAGCTTTATCACCAAAA................................... | 19 | 1 | 2.00 | 2 | 2 | 0 |
| ................................TTGGTGATTTTAGCTGTATGG....................................................................... | 21 | 1 | 2.00 | 2 | 2 | 0 |
| ..............................CTTTGGTGATTTTAGCTGTATGGT...................................................................... | 24 | 1 | 2.00 | 2 | 2 | 0 |
| .................................TGGTGATTTTAGCTGTATG........................................................................ | 19 | 1 | 2.00 | 2 | 2 | 0 |