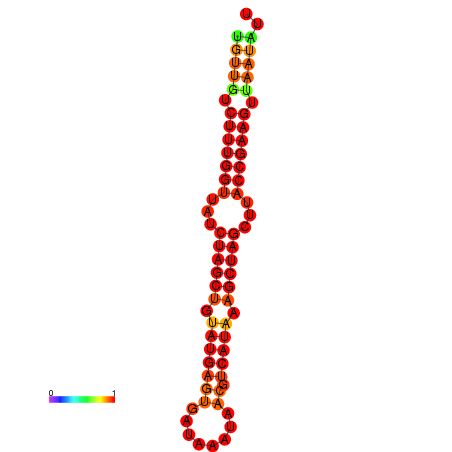
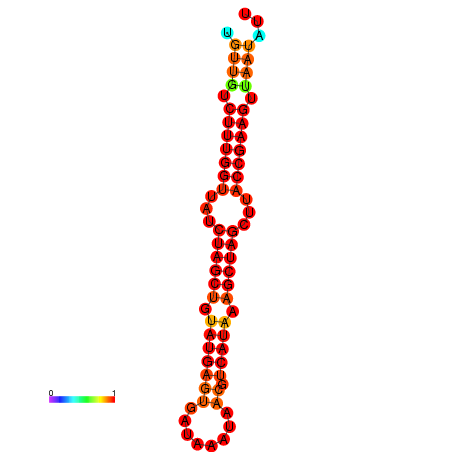
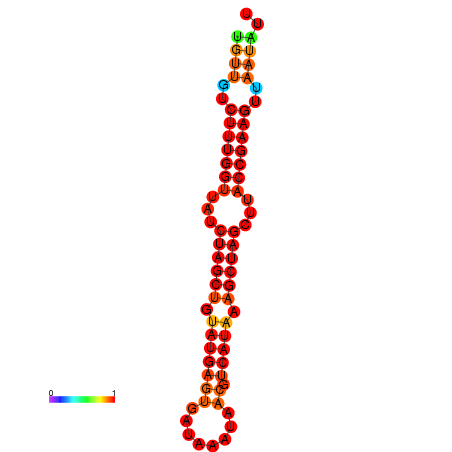
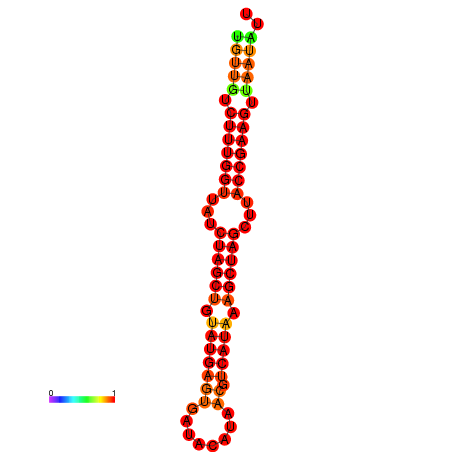
| TTCTTCAATCAGGGTGCTATGTTGTCTTTGGTTATCTAGCTGTATGAGTGATAAATAACGTCATAAAGCTAGCTTACCGAAGTTAATATTAGCGTCTGTCCAGCGAAA | Size | Hit Count | Total Norm | Total | GSM343916
Embryo | GSM444067
Head |
| ........................TCTTTGGTTATCTAGCTGTATG.............................................................. | 22 | 1 | 39978.00 | 39978 | 37618 | 2360 |
| ........................TCTTTGGTTATCTAGCTGTATGA............................................................. | 23 | 1 | 25244.00 | 25244 | 22846 | 2398 |
| ........................TCTTTGGTTATCTAGCTGTAT............................................................... | 21 | 1 | 6922.00 | 6922 | 2520 | 4402 |
| ...............................................................TAAAGCTAGCTTACCGAAGTTA....................... | 22 | 1 | 2318.00 | 2318 | 1801 | 517 |
| ...............................................................TAAAGCTAGCTTACCGAAGTT........................ | 21 | 1 | 767.00 | 767 | 302 | 465 |
| ..............................................................ATAAAGCTAGCTTACCGAAGTTA....................... | 23 | 1 | 608.00 | 608 | 520 | 88 |
| ..............................................................ATAAAGCTAGCTTACCGAAGTT........................ | 22 | 1 | 279.00 | 279 | 180 | 99 |
| ........................TCTTTGGTTATCTAGCTGTA................................................................ | 20 | 1 | 222.00 | 222 | 68 | 154 |
| ...............................................................TAAAGCTAGCTTACCGAAGTTAA...................... | 23 | 1 | 184.00 | 184 | 176 | 8 |
| ........................TCTTTGGTTATCTAGCTGT................................................................. | 19 | 1 | 156.00 | 156 | 15 | 141 |
| ..........................TTTGGTTATCTAGCTGTATG.............................................................. | 20 | 1 | 91.00 | 91 | 84 | 7 |
| ..............................................................ATAAAGCTAGCTTACCGAAGTTAA...................... | 24 | 1 | 69.00 | 69 | 64 | 5 |
| ..........................TTTGGTTATCTAGCTGTATGA............................................................. | 21 | 1 | 62.00 | 62 | 53 | 9 |
| .........................CTTTGGTTATCTAGCTGTATG.............................................................. | 21 | 1 | 30.00 | 30 | 30 | 0 |
| .........................CTTTGGTTATCTAGCTGTATGA............................................................. | 22 | 1 | 28.00 | 28 | 23 | 5 |
| ..............................................................ATAAAGCTAGCTTACCGAAGT......................... | 21 | 1 | 20.00 | 20 | 4 | 16 |
| ...............................................................TAAAGCTAGCTTACCGAAGT......................... | 20 | 1 | 19.00 | 19 | 2 | 17 |
| ..........................TTTGGTTATCTAGCTGTAT............................................................... | 19 | 1 | 14.00 | 14 | 5 | 9 |
| ...........................TTGGTTATCTAGCTGTATGA............................................................. | 20 | 1 | 11.00 | 11 | 9 | 2 |
| ..............................................................ATAAAGCTAGCTTACCGAAG.......................... | 20 | 1 | 9.00 | 9 | 5 | 4 |
| ...........................TTGGTTATCTAGCTGTATG.............................................................. | 19 | 1 | 8.00 | 8 | 8 | 0 |
| ................................................................AAAGCTAGCTTACCGAAGTTA....................... | 21 | 1 | 6.00 | 6 | 5 | 1 |
| ........................TCTTTGGTTATCTAGCTGTATGAG............................................................ | 24 | 1 | 6.00 | 6 | 6 | 0 |