| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3R:17623934-17624068 - | GATG--AAC---GGAG-AAG--------AA--------AGGAT-G-CCGCTCAATTACCCCGACATAGTCATACGGTGAATGTTGTGTATTGGAGACCAATGTTAACTGTAAGACTGTGTCTCGGTGGTTGCCAGCC----CAGCCACA-CAGCCCAGTATCAA |
| droSim1 | chr3R:17429586-17429720 - | GATG--AAC---GGAG-AAG--------AA--------AGGAT-G-CCGCTCAATTACCCCGACATAGTCGTACGGTGAATGTTGTGTATTGGAGACCAATGTTAACTGTAAGACTGTGTCTCGGTGGTTGCCAGCC----CAGCCACA-CAGCCCAGTATCAA |
| droSec1 | super_0:17996302-17996436 - | GATG--AAC---GGAG-AAG--------AA--------AGGAT-G-CCGCTCAATTACCCCGACATAGTCATACGGTGAATGTTGTGTATTGGAGACCAATGTTAACTGTAAGACTGTGTCTCGGTGGTTGCCAGCC----CAGCCACA-CAGCCCAGTATCAA |
| droYak2 | chr3R:18443923-18444058 - | AA-----AT---CGAG-AAG--------AA--------AGGAT-G-CCGCTCAATTACCCCGACATAGTCATACGGTGAATGTTGTGTATTGGAGACCAATGTTAACTGTAAGACTGTGTCTCGGTGGTTGCCAGCCACACCAGCCACATCAGCC--ACATCGG |
| droEre2 | scaffold_4820:10463202-10463335 + | GATG--AAC---GAAG-AAG--------AA--------AGGAT-G-CCGCTCAATTACCCCGACATAGTCATACGGTGAATGTTGTGTATTGGAGACCAATGTTAACTGTAAGACTGTGTCTCGGTGGTTGCCAGCC----CAGCCACACCAGCC--ACACCAG |
| droAna3 | scaffold_13340:8622498-8622632 - | GTTGCCAAA---GGACCAAA--------AA--------AGGAT-G-CCGCTCAATTACCCCGACATAGTCGTACGGTGAATGTTGTGTATTGGAAAGCAATGTTAACTGTAAGACTGTGTCTCGGTGGTTGCCAGCT-----AGCCTAC-CATCTCACTATC-- |
| dp4 | chr2:6480711-6480794 - | ---------------------------------------------------------------CATAGTCGTACGGTGAATGTTGTGTATTGG--ACCAATGTTAACTGTAAGACTGTGTCTCGGTGGTTGTCAGCC----AAGCAGCA--AGCC--------- |
| droPer1 | super_0:306015-306135 - | GATG----TGAAGGAG-CCC--------AA--------AGGAT-GCCCGCTCAATTACCATGACATAGTCGTACGGTGAATGTTGTGTATTGG--ACCAATGTTAACTGTAAGACTGTGTCTCGGTGGTTGTCAGCC----AAGCAGCA--------------- |
| droWil1 | scaffold_181130:11672712-11672848 - | GACG--A-C---GA-CGACGACGCCAACGACAGCAGCAAAGATGTCCCGCTCAATTACCATGACATAGTCGTACGGTGAATGTTGTTTA-TGGAGTCCAATGTTAACTGTAAGACTGTGTCTCGGTGGTTGCCAGCC----AAACAACA--------------- |
| droVir3 | scaffold_12822:3936742-3936840 - | G---------------------------------------------CCGCTCAATTACCGTGACATAGTCGTACGGTGAATGTTGTGTA-TGGAGTCCAATGTTAACTGTAAGACTGTGTCTCGGTGGTTGTTAGCT----AATCAACA--------------- |
| droMoj3 | scaffold_6540:17209641-17209764 - | ATTA--AAA---GG-G-CAG--------TG--------A-GTC-G-CCGCTCAATTACCGTGACATAGTCGTACGGTGAATGTTGTTTA-TGGAGTCCAATGTTAACTGTAAGACTGTGTCTCGGTGGTTG-CAGCC----AAACAACA-GCGACC-------- |
| droGri2 | scaffold_8129:786-906 - | GATA----A---GGACAAGA--------GAC-------A-GCA-G-TCGCTCAATTACCATGACATAGTCGTACGGTGAATGTTGTGT-TTGGAGTCCAATGTTAACTGTAAGACTGTGTCTTGGTGGTTGCCAGCC----AAGCAACA-AA------------ |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr3R:17623934-17624068 - | GATG--AAC---GGAG-AAG--------AA--------AGGAT-G-CCGCTCAATTACCCCGACATAGTCATACGGTGAATGTTGTGTATTGGAGACCAATGTTAACTGTAAGACTGTGTCTCGGTGGTTGCCAGCC----CAGCCACA-CAGCCCAGTATCAA |
| droSim1 | chr3R:17429586-17429720 - | GATG--AAC---GGAG-AAG--------AA--------AGGAT-G-CCGCTCAATTACCCCGACATAGTCGTACGGTGAATGTTGTGTATTGGAGACCAATGTTAACTGTAAGACTGTGTCTCGGTGGTTGCCAGCC----CAGCCACA-CAGCCCAGTATCAA |
| droSec1 | super_0:17996302-17996436 - | GATG--AAC---GGAG-AAG--------AA--------AGGAT-G-CCGCTCAATTACCCCGACATAGTCATACGGTGAATGTTGTGTATTGGAGACCAATGTTAACTGTAAGACTGTGTCTCGGTGGTTGCCAGCC----CAGCCACA-CAGCCCAGTATCAA |
| droYak2 | chr3R:18443923-18444058 - | AA-----AT---CGAG-AAG--------AA--------AGGAT-G-CCGCTCAATTACCCCGACATAGTCATACGGTGAATGTTGTGTATTGGAGACCAATGTTAACTGTAAGACTGTGTCTCGGTGGTTGCCAGCCACACCAGCCACATCAGCC--ACATCGG |
| droEre2 | scaffold_4820:10463202-10463335 + | GATG--AAC---GAAG-AAG--------AA--------AGGAT-G-CCGCTCAATTACCCCGACATAGTCATACGGTGAATGTTGTGTATTGGAGACCAATGTTAACTGTAAGACTGTGTCTCGGTGGTTGCCAGCC----CAGCCACACCAGCC--ACACCAG |
| droAna3 | scaffold_13340:8622498-8622632 - | GTTGCCAAA---GGACCAAA--------AA--------AGGAT-G-CCGCTCAATTACCCCGACATAGTCGTACGGTGAATGTTGTGTATTGGAAAGCAATGTTAACTGTAAGACTGTGTCTCGGTGGTTGCCAGCT-----AGCCTAC-CATCTCACTATC-- |
| dp4 | chr2:6480711-6480794 - | ---------------------------------------------------------------CATAGTCGTACGGTGAATGTTGTGTATTGG--ACCAATGTTAACTGTAAGACTGTGTCTCGGTGGTTGTCAGCC----AAGCAGCA--AGCC--------- |
| droPer1 | super_0:306015-306135 - | GATG----TGAAGGAG-CCC--------AA--------AGGAT-GCCCGCTCAATTACCATGACATAGTCGTACGGTGAATGTTGTGTATTGG--ACCAATGTTAACTGTAAGACTGTGTCTCGGTGGTTGTCAGCC----AAGCAGCA--------------- |
| droWil1 | scaffold_181130:11672712-11672848 - | GACG--A-C---GA-CGACGACGCCAACGACAGCAGCAAAGATGTCCCGCTCAATTACCATGACATAGTCGTACGGTGAATGTTGTTTA-TGGAGTCCAATGTTAACTGTAAGACTGTGTCTCGGTGGTTGCCAGCC----AAACAACA--------------- |
| droVir3 | scaffold_12822:3936742-3936840 - | G---------------------------------------------CCGCTCAATTACCGTGACATAGTCGTACGGTGAATGTTGTGTA-TGGAGTCCAATGTTAACTGTAAGACTGTGTCTCGGTGGTTGTTAGCT----AATCAACA--------------- |
| droMoj3 | scaffold_6540:17209641-17209764 - | ATTA--AAA---GG-G-CAG--------TG--------A-GTC-G-CCGCTCAATTACCGTGACATAGTCGTACGGTGAATGTTGTTTA-TGGAGTCCAATGTTAACTGTAAGACTGTGTCTCGGTGGTTG-CAGCC----AAACAACA-GCGACC-------- |
| droGri2 | scaffold_8129:786-906 - | GATA----A---GGACAAGA--------GAC-------A-GCA-G-TCGCTCAATTACCATGACATAGTCGTACGGTGAATGTTGTGT-TTGGAGTCCAATGTTAACTGTAAGACTGTGTCTTGGTGGTTGCCAGCC----AAGCAACA-AA------------ |
| Species | Read pileup | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
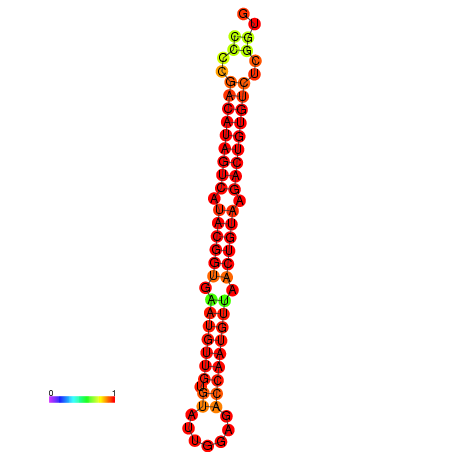
| dG=-27.5, p-value=0.009901 | dG=-27.2, p-value=0.009901 |
|---|---|
 |
 |
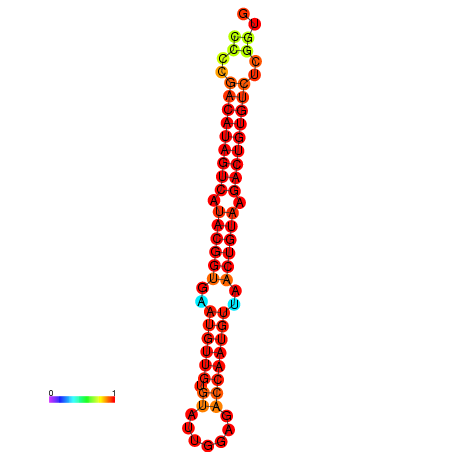
| dG=-27.5, p-value=0.009901 | dG=-27.2, p-value=0.009901 |
|---|---|
 |
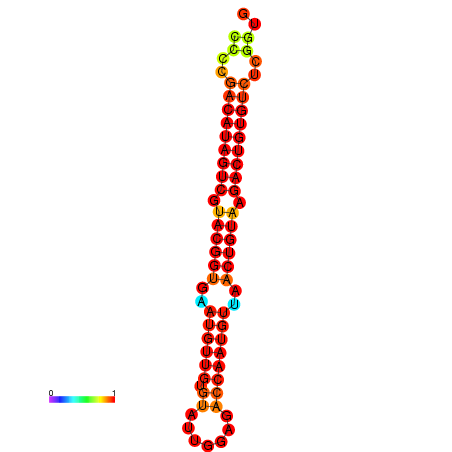 |
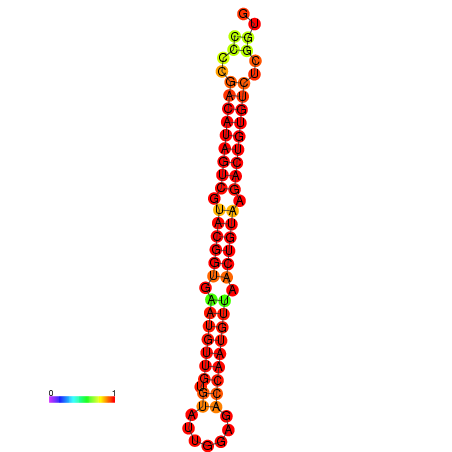
| dG=-27.5, p-value=0.009901 | dG=-27.2, p-value=0.009901 |
|---|---|
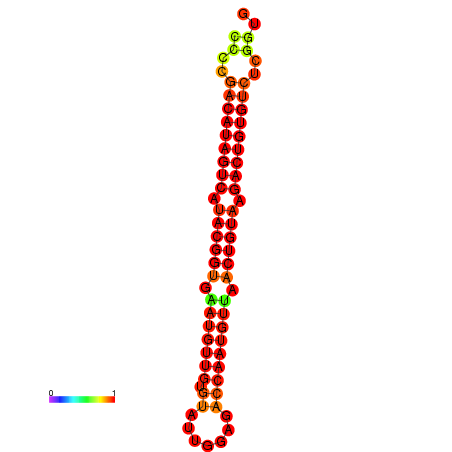 |
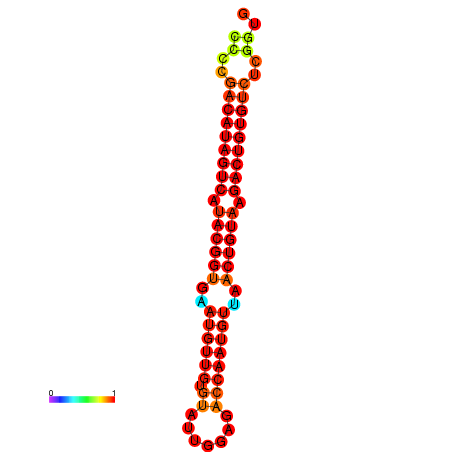 |
| dG=-27.5, p-value=0.009901 | dG=-27.2, p-value=0.009901 |
|---|---|
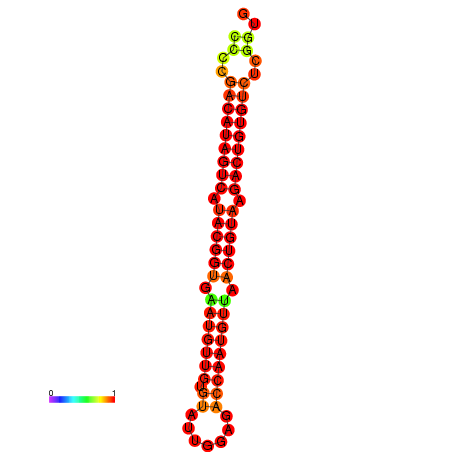 |
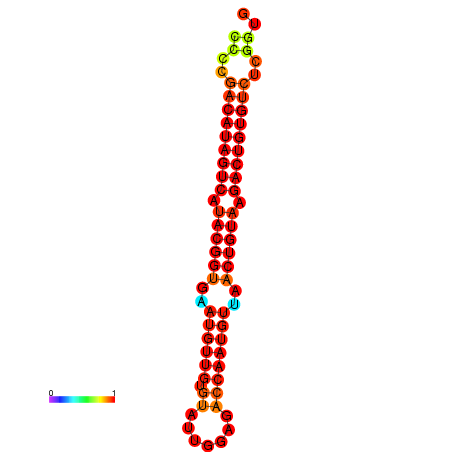 |
| dG=-27.5, p-value=0.009901 | dG=-27.2, p-value=0.009901 |
|---|---|
 |
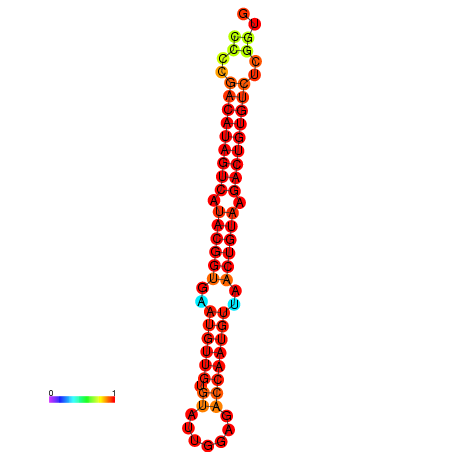 |
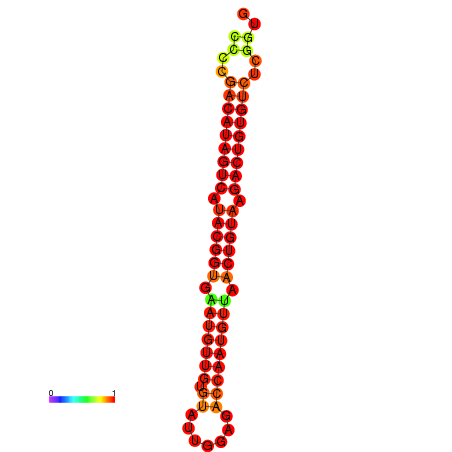
| dG=-28.2, p-value=0.009901 | dG=-27.9, p-value=0.009901 |
|---|---|
 |
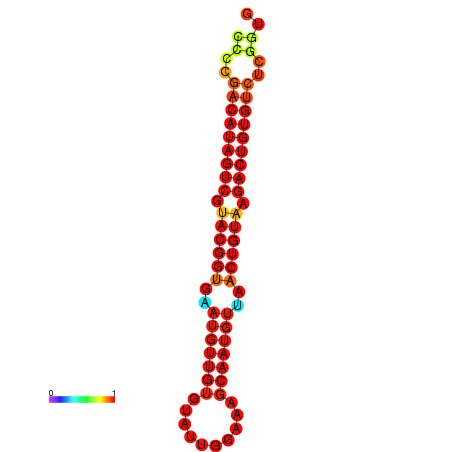 |
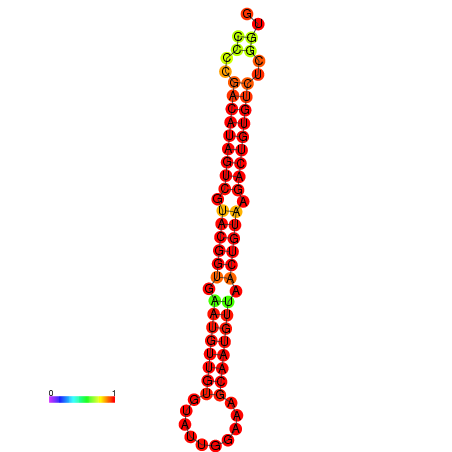
| dG=-21.4, p-value=0.009901 | dG=-21.1, p-value=0.009901 |
|---|---|
 |
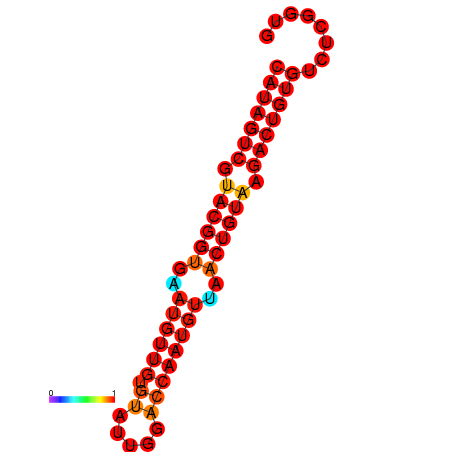 |
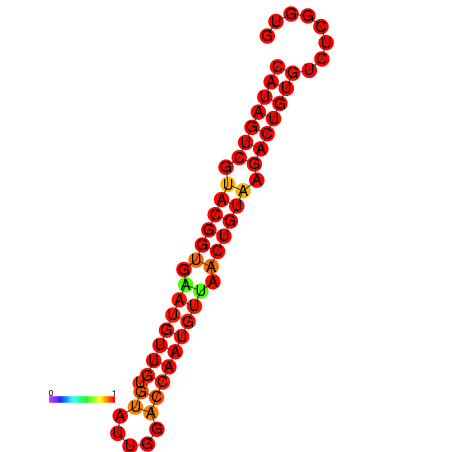
| dG=-28.5, p-value=0.009901 | dG=-28.2, p-value=0.009901 |
|---|---|
 |
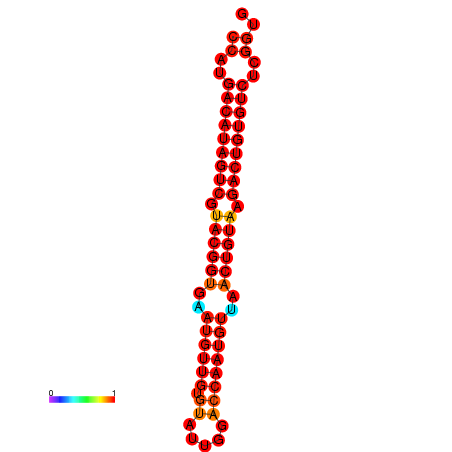 |
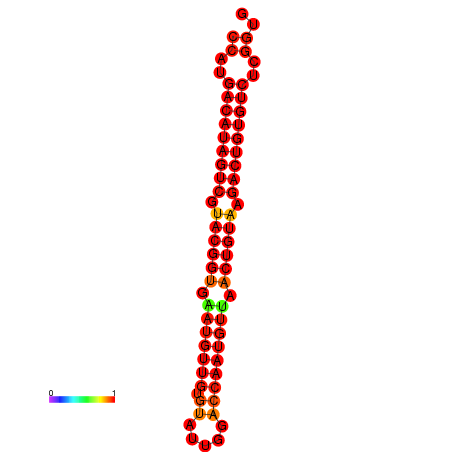
| dG=-26.4, p-value=0.009901 | dG=-26.1, p-value=0.009901 |
|---|---|
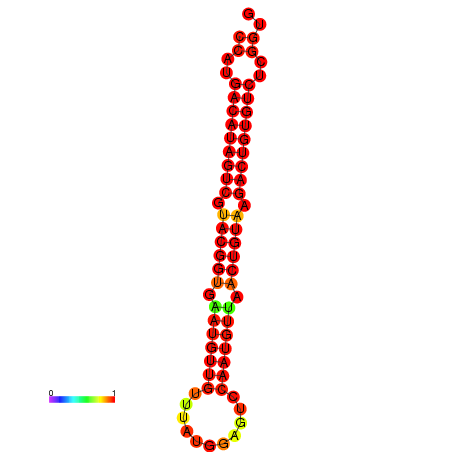 |
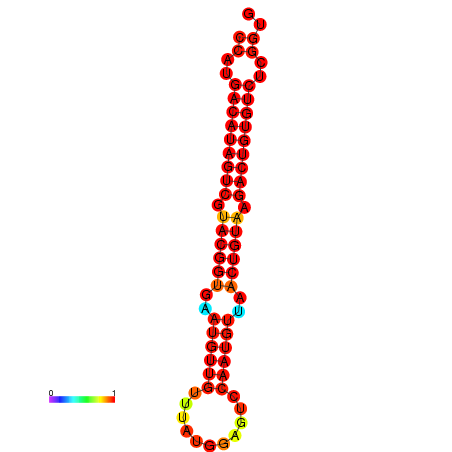 |
| dG=-29.7, p-value=0.009901 | dG=-29.4, p-value=0.009901 |
|---|---|
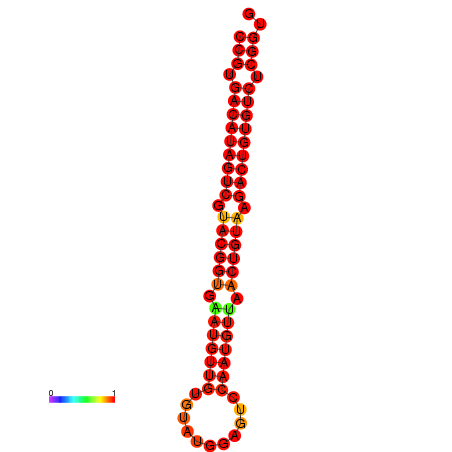 |
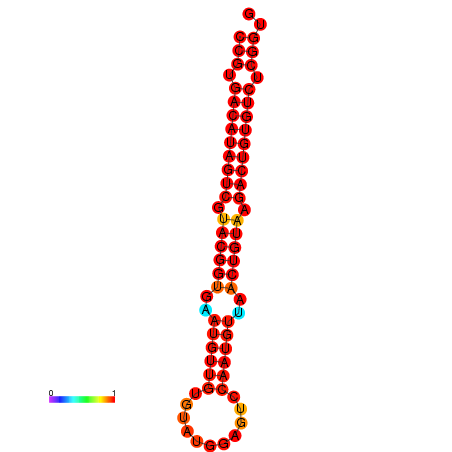 |
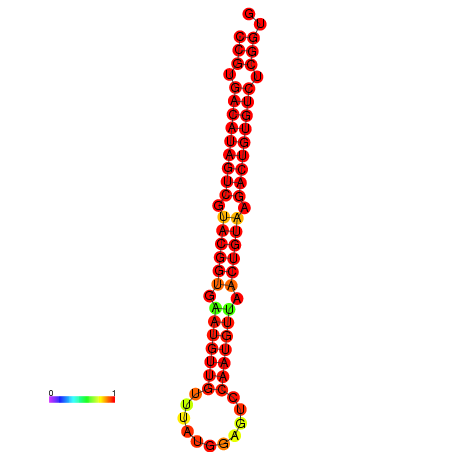
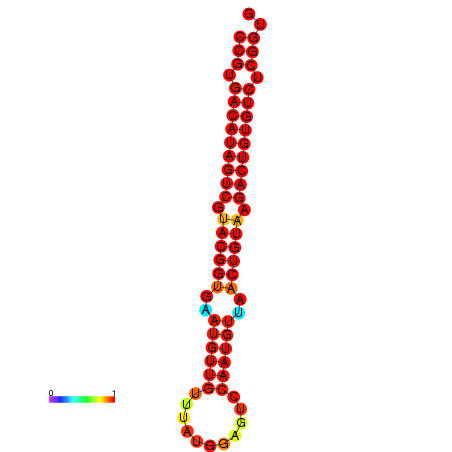
| dG=-29.7, p-value=0.009901 | dG=-29.4, p-value=0.009901 |
|---|---|
 |
 |
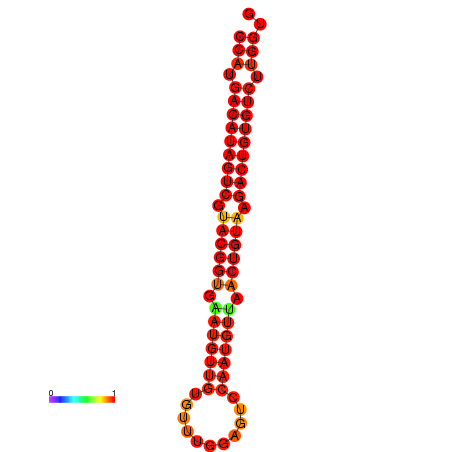
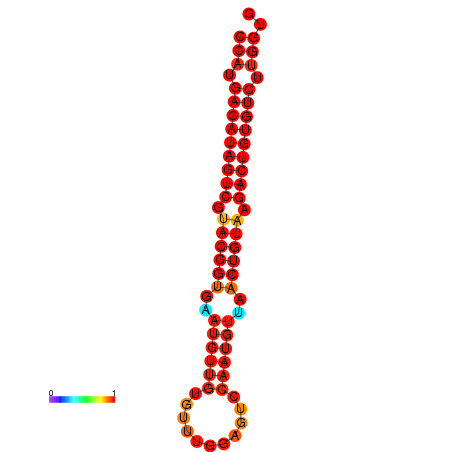
| dG=-28.5, p-value=0.009901 | dG=-28.2, p-value=0.009901 |
|---|---|
 |
 |
Generated: 03/07/2013 at 07:27 PM