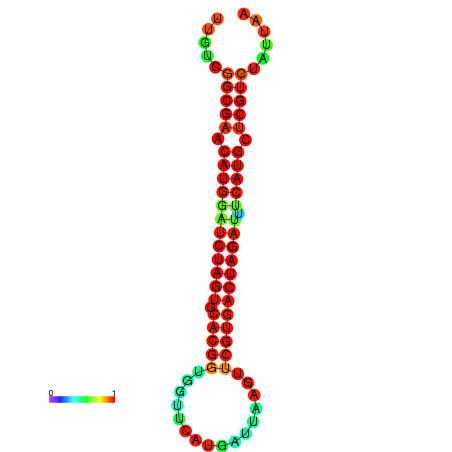
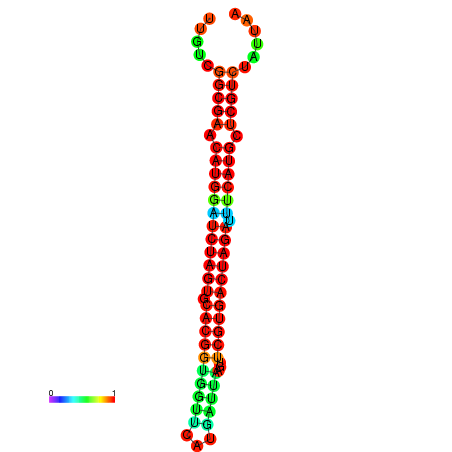
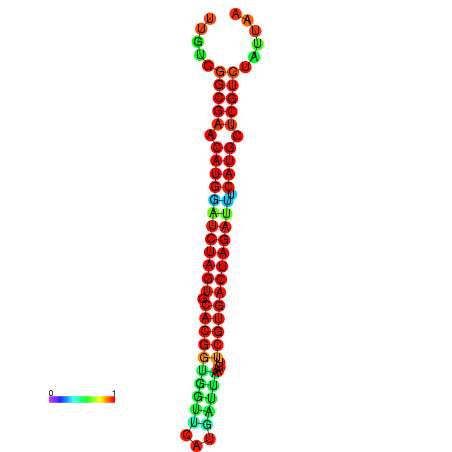
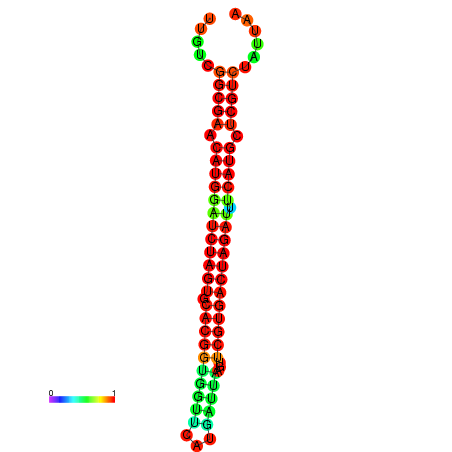
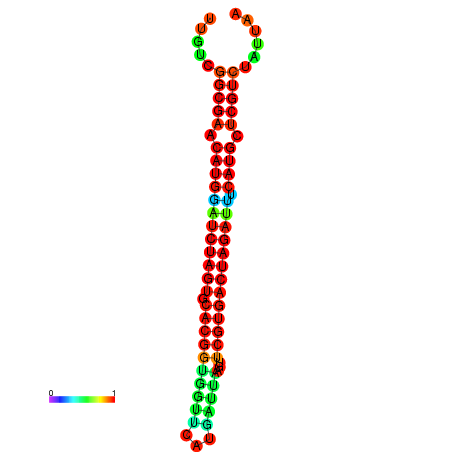
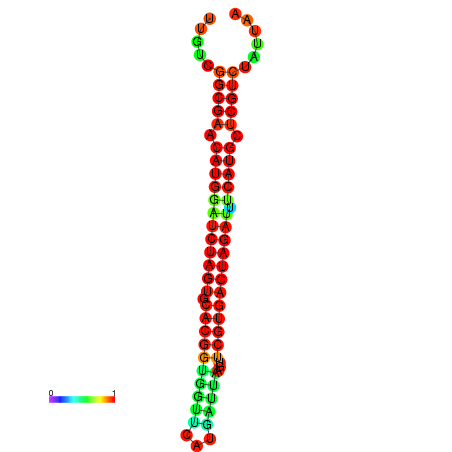
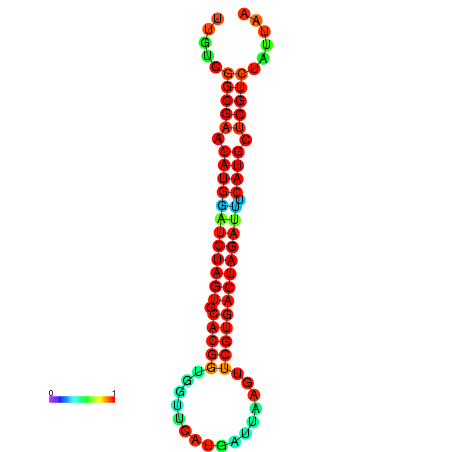
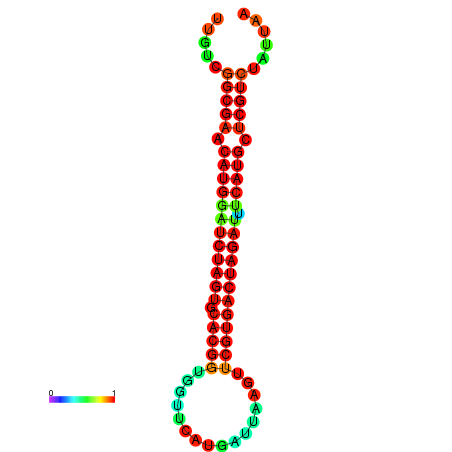
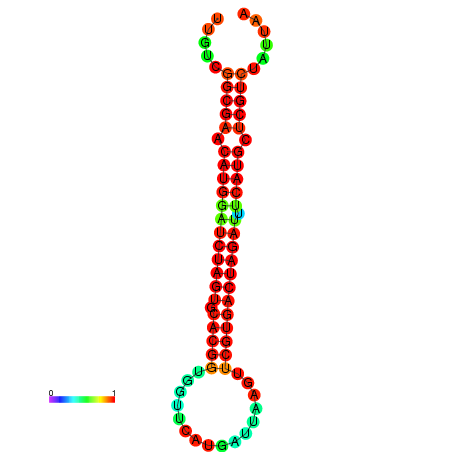
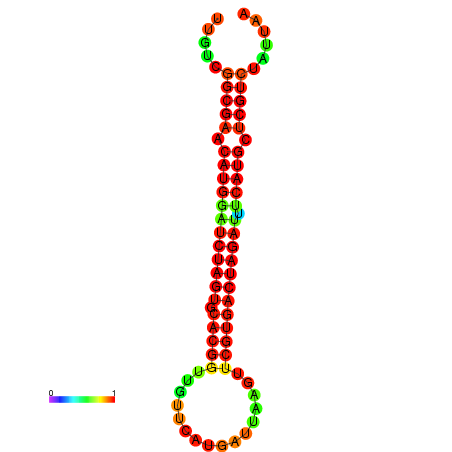
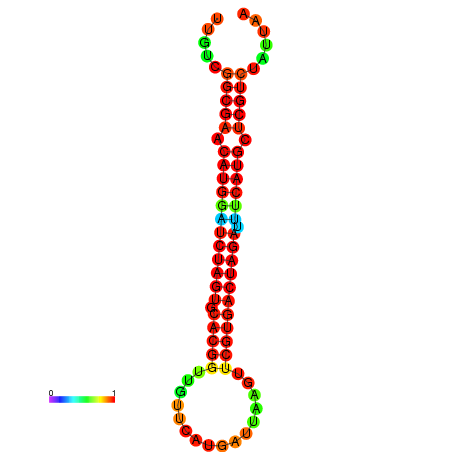
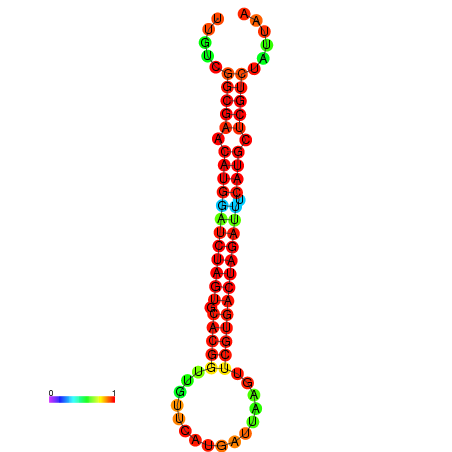
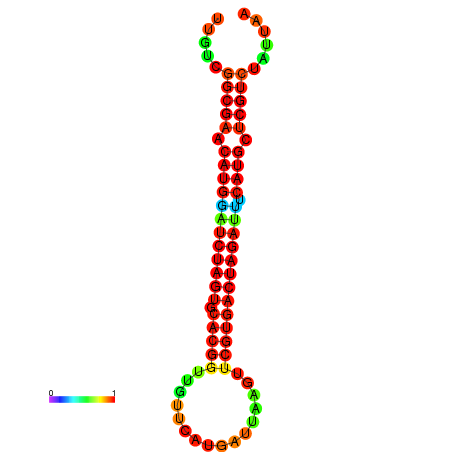
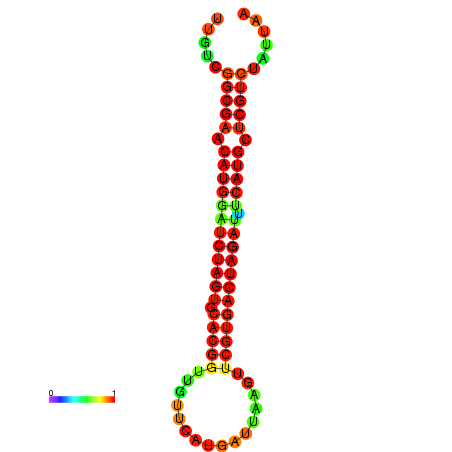
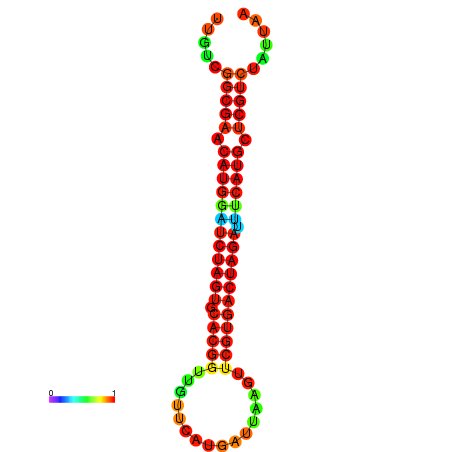
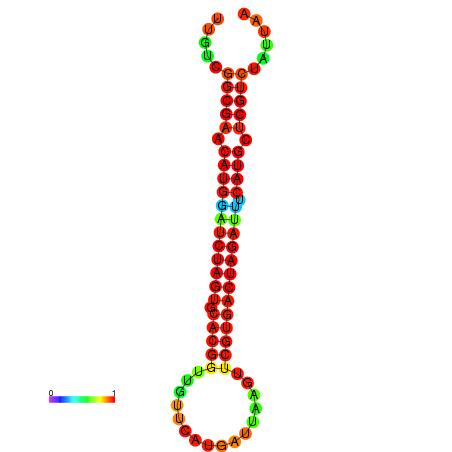
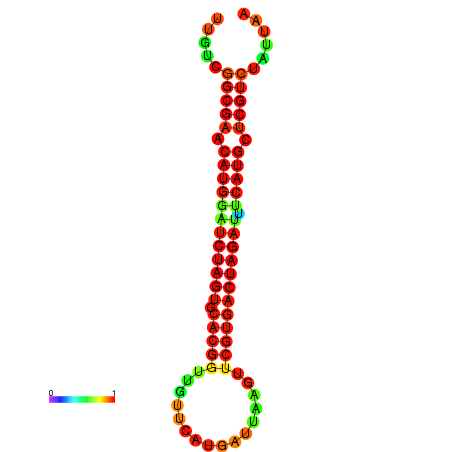
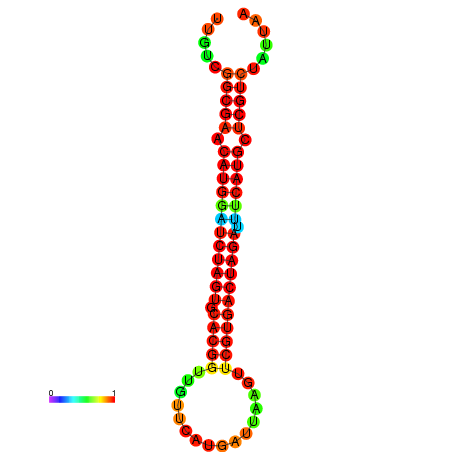
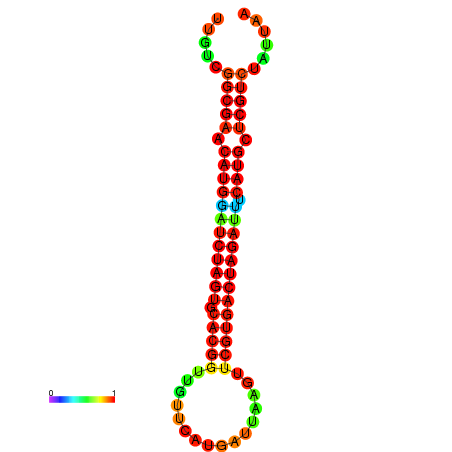
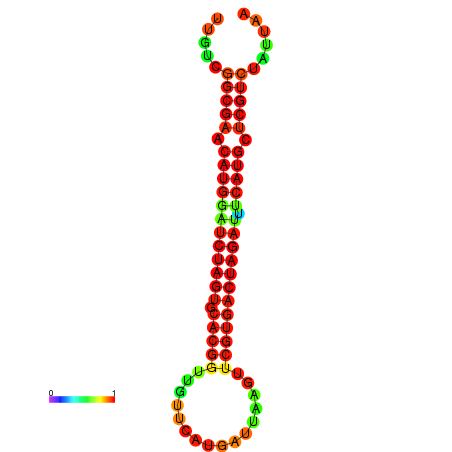
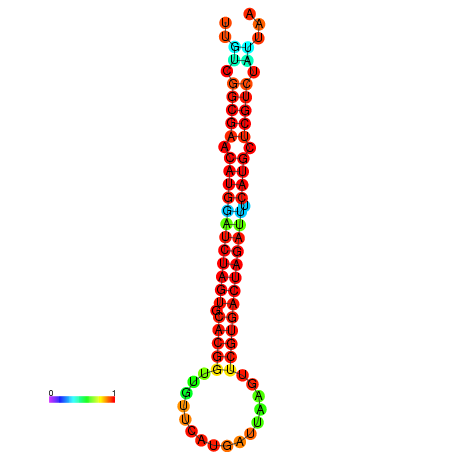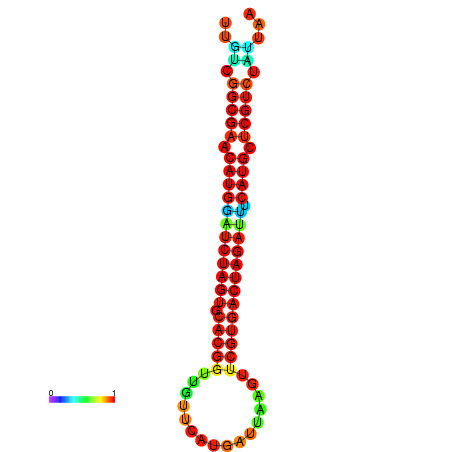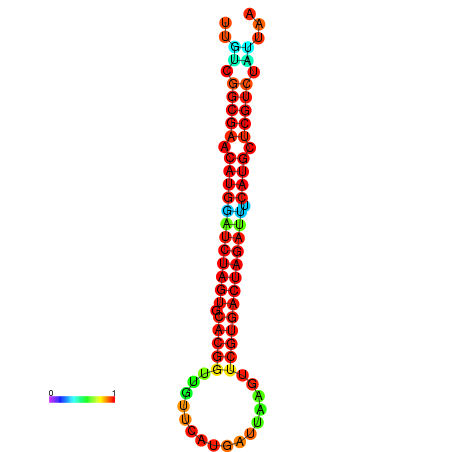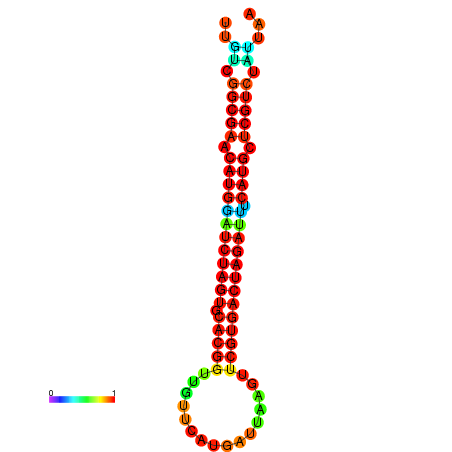| GGAAGGATGCTCTTTTCTGACTCTATTTTGTCGGCGAACATGGATCTAGTGCACGGTGGTTCATGATTAAGTTCGTGACTAGATTTCATGCTCGTCTATTAAGTTGGGTCAGCACAACGAA----GAGAGC | Size | Hit Count | Total Norm | Total | GSM343915
Embryo |
| ...........................................................................TGACTAGATTTCATGCTCGTCT.................................. | 22 | 1 | 922.00 | 922 | 922 |
| ...........................................................................TGACTAGATTTCATGCTCGTCTA................................. | 23 | 1 | 814.00 | 814 | 814 |
| ...........................................................................TGACTAGATTTCATGCTCGT.................................... | 20 | 1 | 401.00 | 401 | 401 |
| .................................GCGAACATGGATCTAGTGCACG............................................................................ | 22 | 1 | 312.00 | 312 | 312 |
| ................................GGCGAACATGGATCTAGTGCACG............................................................................ | 23 | 1 | 309.00 | 309 | 309 |
| ...........................................................................TGACTAGATTTCATGCTCGTC................................... | 21 | 1 | 216.00 | 216 | 216 |
| ..........................................................................GTGACTAGATTTCATGCTCGTCT.................................. | 23 | 1 | 50.00 | 50 | 50 |
| .......................................................GTGGTTCATGATTAAGTTCG........................................................ | 20 | 1 | 31.00 | 31 | 31 |
| .................................GCGAACATGGATCTAGTGCACGG........................................................................... | 23 | 1 | 19.00 | 19 | 19 |
| ................................GGCGAACATGGATCTAGTGCA.............................................................................. | 21 | 1 | 18.00 | 18 | 18 |
| ................................GGCGAACATGGATCTAGTGCAC............................................................................. | 22 | 1 | 15.00 | 15 | 15 |
| ................................GGCGAACATGGATCTAGTGC............................................................................... | 20 | 1 | 14.00 | 14 | 14 |
| ..................................CGAACATGGATCTAGTGCACGG........................................................................... | 22 | 1 | 13.00 | 13 | 13 |
| ..........................................................................GTGACTAGATTTCATGCTCGTC................................... | 22 | 1 | 10.00 | 10 | 10 |
| .................................GCGAACATGGATCTAGTGCAC............................................................................. | 21 | 1 | 7.00 | 7 | 7 |
| ..........................................................................GTGACTAGATTTCATGCTCGT.................................... | 21 | 1 | 6.00 | 6 | 6 |
| ...............................CGGCGAACATGGATCTAGTGCACG............................................................................ | 24 | 1 | 5.00 | 5 | 5 |
| .................................GCGAACATGGATCTAGTGC............................................................................... | 19 | 1 | 4.00 | 4 | 4 |
| .............................................................................ACTAGATTTCATGCTCGTCT.................................. | 20 | 1 | 4.00 | 4 | 4 |
| .............................................................................ACTAGATTTCATGCTCGTCTA................................. | 21 | 1 | 3.00 | 3 | 3 |
| .................................GCGAACATGGATCTAGTGCA.............................................................................. | 20 | 1 | 3.00 | 3 | 3 |
| .................................GCGAACATGGATCTAGTGCACGGT.......................................................................... | 24 | 1 | 3.00 | 3 | 3 |
| ...........................................................................TGACTAGATTTCATGCTC...................................... | 18 | 1 | 2.00 | 2 | 2 |
| ...........................................................................TGACTAGATTTCATGCTCGTCTAT................................ | 24 | 1 | 2.00 | 2 | 2 |
| ............................................................................GACTAGATTTCATGCTCGT.................................... | 19 | 1 | 2.00 | 2 | 2 |
| ...................................GAACATGGATCTAGTGCACG............................................................................ | 20 | 1 | 2.00 | 2 | 2 |
| ..................................CGAACATGGATCTAGTGCACG............................................................................ | 21 | 1 | 1.00 | 1 | 1 |
| ............................................................................GACTAGATTTCATGCTCGTC................................... | 20 | 1 | 1.00 | 1 | 1 |
| ...........................................................................TGACTAGATTTCATGCTCG..................................... | 19 | 1 | 1.00 | 1 | 1 |
| .............................................................................ACTAGATTTCATGCTCGTC................................... | 19 | 1 | 1.00 | 1 | 1 |
| ................................GGCGAACATGGATCTAGTGCACGG........................................................................... | 24 | 1 | 1.00 | 1 | 1 |