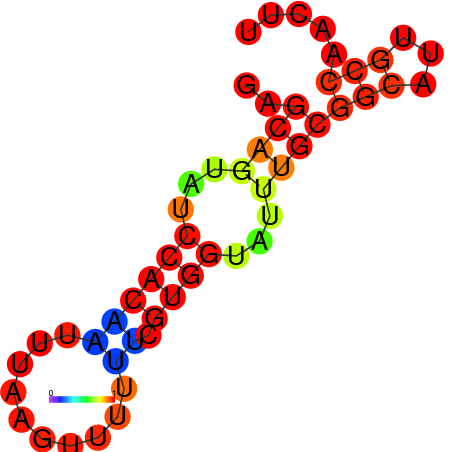
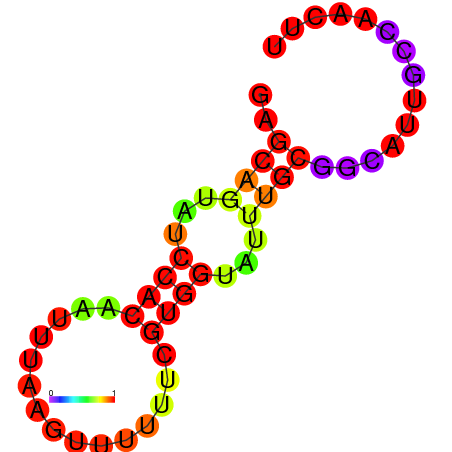
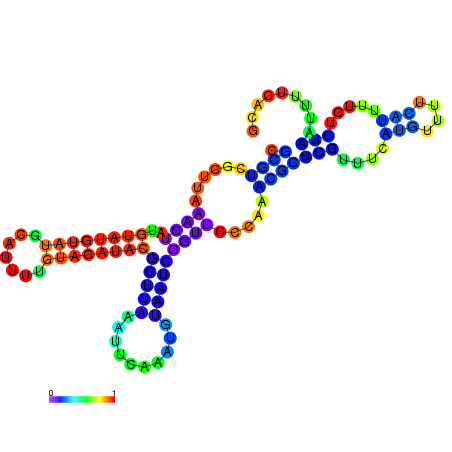
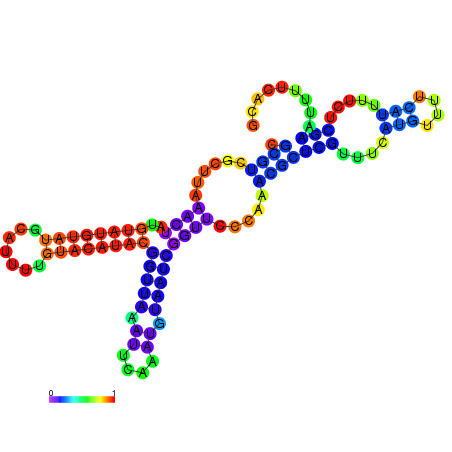
| dm3 |
chr2R:16472804-16472925 - |
ACAAAT--------CT-----AGACATGATTTTCCCAAGTGCCTGGTATCAGCAAAGTGT-----TATTTTTTATGTTT-------------------------------------------------ATGTAAAGTACACGTTTCTGGTACTAAGTACTTCGAGAAAGTT---------------ACCTGCGTAGGATATCAA |
| droSim1 |
chr2R:15134498-15134613 - |
AGGAAT--------CT-----AGACATGATTTTCGCAAGTGCCTGGTTTCAGAGAGGTGT-----AATTTTT---G----------------------------------------------------TTGTATAGTACCCTTTTTTGGAACCAAGCCCTTTGAGAAACTT---------------ACCTGCGGGGTATATCAA |
| droSec1 |
super_1:14027523-14027641 - |
AGGAAT--------CT-----AGACATGATTTTCGGAAGTGCCTGGTATCAGATAGGTGT-----AATTTTT---GTGT-------------------------------------------------TTGTATAGTACACGTTTCTGGAACCAAGCCCTTTGAGAAACTT---------------ACCTGCGGGGTATATCAA |
| droYak2 |
chr2R:12723658-12723777 + |
AGGACT--------CT--T--GGATCTGATTTTCCCAAGTGCTTAGTATCAGCAAGGTGT-----AATTTTT---GTTT-------------------------------------------------GGTTAAAGTACACGTTACTGGTACTACGTACTTCGAGAAACCT---------------AACTACGGAGGATACAAA |
| droEre2 |
scaffold_4845:10619147-10619257 - |
AGAAC--------------------CTGATTTTCCCAAGTGCATAGTATCAGCAAGGTGT-----AATTTTT---ATTT-------------------------------------------------GGGTAAAGTACACGTTACTGGTACTAGGTACTTTGAGAAACAT---------------GACTACGGAGAATACCA- |
| droAna3 |
scaffold_13266:4131420-4131545 + |
TAAAAT--------TCATTGTAAACCATACTTTTTAAG--GAGCAGTATCCACAATTTAA---------------GTTT-------------------------------------------------TTTCGTGGT----ATTTGCGGCATTGCCAACTTTAAGAAAAGTTCAAAGGGCGCTGTTACAAGCTATTACTATTAC |
| dp4 |
chr3:8679002-8679138 + |
GTCGTT--------CT-----CCATCTCTTTCTCGCGTC-GCGTCGCTTAAACTATGTATGTATGCATTTTGTACATACGGTTAAATTCAAATGTA-------------ATCGGTTCCCAAACGCTCGTTTCATGTTTTCATTTTCTCGAATTTTCACG----------------------------------------TTTAG |
| droPer1 |
Unknown |
------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ |
| droWil1 |
scaffold_181096:3840730-3840836 + |
GCATT---------AT-----AGAAAACA--------AACGATTGTT----------------------------------------TACTGGCCATTTTGCTAATCTTTT-----------------TGGTAATCTAATC-TTTACGGTATCAAAATTAGTTAGAAAA-T---------------ACTTAATAATTATATTA- |
| droVir3 |
scaffold_13042:3503607-3503688 - |
ATAAAAGGCAATTCCT--T--AGAATT--TTTCTTCAATTAATTAATTCT--------GT-----TATGTTTTATATT-------------------------------------------------------------TAATTTCCAATAATAAGTTCTTC------------------------------------------ |
| droMoj3 |
scaffold_6680:4944926-4945004 - |
GTTTAT--------AT---------ATG-------AAAATATTTGATATATGCAA------------TTTAT--------------ACAAAGAACTTTTTGCTA-------------------------------------------------------TTCTAAAAAATA---------------AA------CGAACATCCA |
| droGri2 |
Unknown |
------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ |