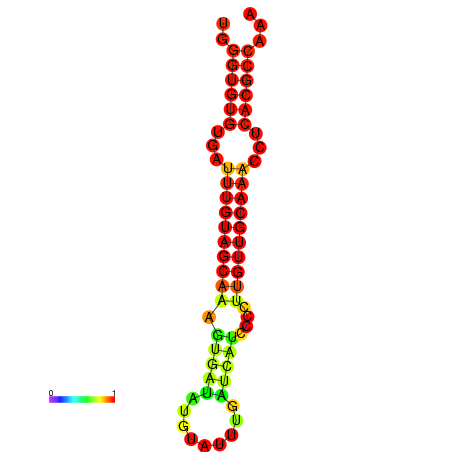
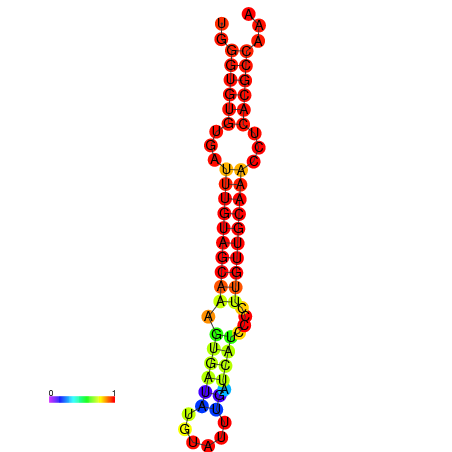
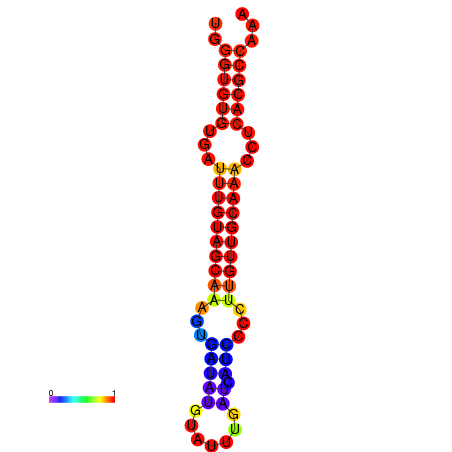
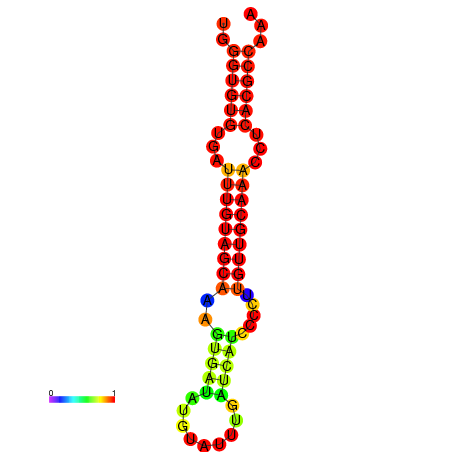
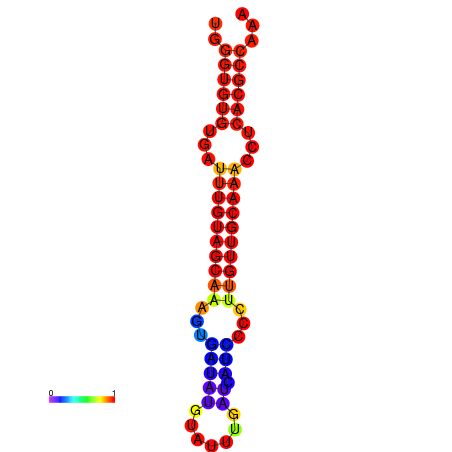
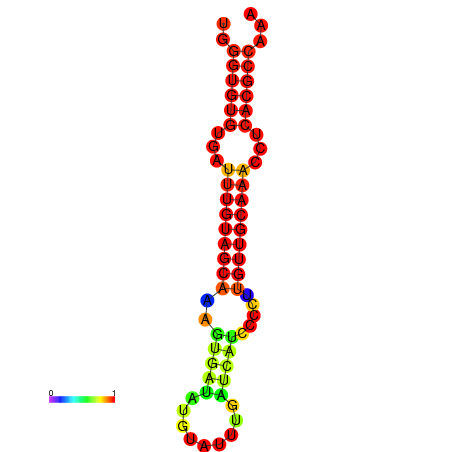
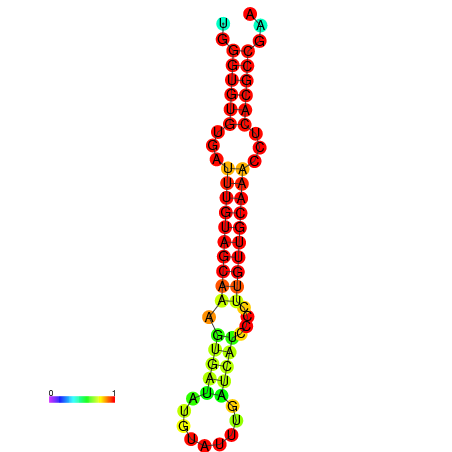
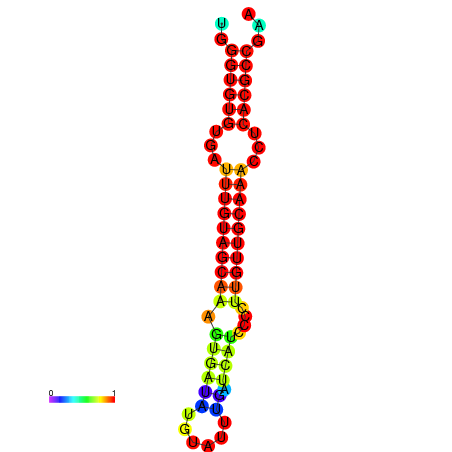
| CCGGATAACTG----------GGTATCCAGGAATCCAGACTGTGGACATTTTGGGTGTGTGATTTGTGGCAAAGTGATATGCATTTGATCATCCCCTTGTTGCAAACCTCACGCCAAAGTTGAATCCCACACCTACACTCTCATATAATGATCAT | Size | Hit Count | Total Norm | Total | GSM343916
Embryo | GSM444067
Head |
| ............................................................................................CCCCTTGTTGCAAACCTCACG.......................................... | 21 | 1 | 456.00 | 456 | 290 | 166 |
| ............................................................................................CCCCTTGTTGCAAACCTCACGC......................................... | 22 | 1 | 420.00 | 420 | 199 | 221 |
| ........................................................GTGTGATTTGTGGCAAAGTGAT............................................................................. | 22 | 1 | 45.00 | 45 | 17 | 28 |
| ........................................................GTGTGATTTGTGGCAAAGTG............................................................................... | 20 | 1 | 38.00 | 38 | 3 | 35 |
| .........................................................TGTGATTTGTGGCAAAGTGAT............................................................................. | 21 | 1 | 27.00 | 27 | 5 | 22 |
| ........................................................GTGTGATTTGTGGCAAAGTGA.............................................................................. | 21 | 1 | 24.00 | 24 | 6 | 18 |
| .........................................................TGTGATTTGTGGCAAAGTGA.............................................................................. | 20 | 1 | 16.00 | 16 | 0 | 16 |
| .........................................................TGTGATTTGTGGCAAAGTGATA............................................................................ | 22 | 1 | 10.00 | 10 | 3 | 7 |
| ............................................................................................CCCCTTGTTGCAAACCTCAC........................................... | 20 | 1 | 8.00 | 8 | 5 | 3 |
| .............................................................................................CCCTTGTTGCAAACCTCACG.......................................... | 20 | 1 | 8.00 | 8 | 6 | 2 |
| .............................................................................................CCCTTGTTGCAAACCTCACGC......................................... | 21 | 1 | 7.00 | 7 | 2 | 5 |
| ........................................................GTGTGATTTGTGGCAAAGT................................................................................ | 19 | 1 | 7.00 | 7 | 1 | 6 |
| ...........................................................................................TCCCCTTGTTGCAAACCTCACG.......................................... | 22 | 1 | 6.00 | 6 | 4 | 2 |
| .......................................................TGTGTGATTTGTGGCAAAGTG............................................................................... | 21 | 1 | 4.00 | 4 | 0 | 4 |
| .....................................GACTGTGGACATTTTGGGT................................................................................................... | 19 | 1 | 4.00 | 4 | 4 | 0 |
| ........................................................GTGTGATTTGTGGCAAAG................................................................................. | 18 | 1 | 3.00 | 3 | 1 | 2 |
| .......................................................TGTGTGATTTGTGGCAAAGTGAT............................................................................. | 23 | 1 | 3.00 | 3 | 2 | 1 |
| ...............................................................................................CTTGTTGCAAACCTCACGC......................................... | 19 | 1 | 3.00 | 3 | 3 | 0 |
| .......................................................TGTGTGATTTGTGGCAAAGTGA.............................................................................. | 22 | 1 | 2.00 | 2 | 0 | 2 |
| ............................................................................................CCCCTTGTTGCAAACCTCA............................................ | 19 | 1 | 2.00 | 2 | 1 | 1 |
| ............................................................................................CCCCTTGTTGCAAACCTCACGCC........................................ | 23 | 1 | 1.00 | 1 | 1 | 0 |
| ........................................................GTGTGATTTGTGGCAAAGTGATA............................................................................ | 23 | 1 | 1.00 | 1 | 0 | 1 |
| .......................................................TGTGTGATTTGTGGCAAAG................................................................................. | 19 | 1 | 1.00 | 1 | 0 | 1 |
| ................................................................................................TTGTTGCAAACCTCACGC......................................... | 18 | 1 | 1.00 | 1 | 1 | 0 |
| ...............................................................................................CTTGTTGCAAACCTCACG.......................................... | 18 | 1 | 1.00 | 1 | 1 | 0 |
| ...........................................................TGATTTGTGGCAAAGTGAT............................................................................. | 19 | 1 | 1.00 | 1 | 0 | 1 |