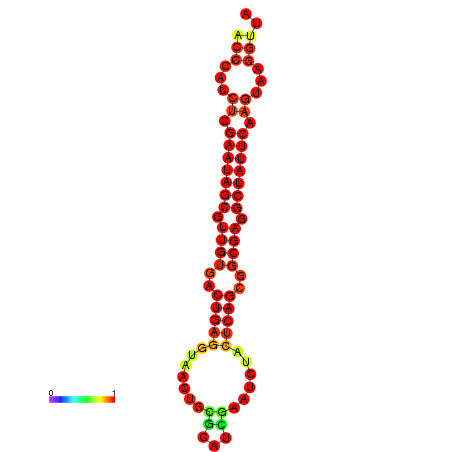

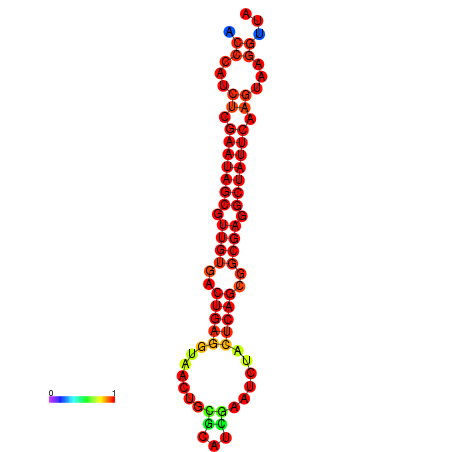

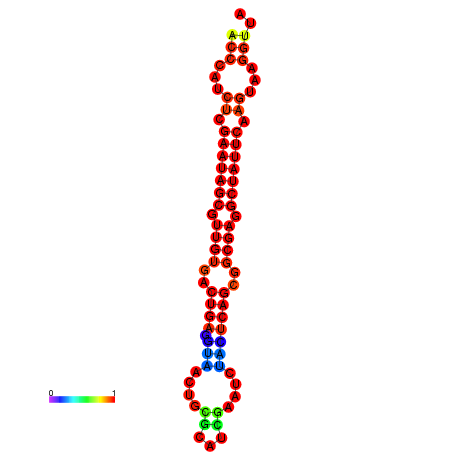
| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2R:4332472-4332601 + | TAGCCACGCAG--AGGA---------CACACCTG-------AAATTACCCATCTCGAATAGCGTTGTGACTGA-GGTAACTGCGCATCGAATCTACTCAGCGGCGAGGCTATTCAAGTAAGGTTATTTTGGGC----CAGCGACTTTA--CGACC |
| droSim1 | chr2R:2956848-2956977 + | TAGCCACGCAG--AGGA---------CACACCTG-------AAATTACCCATCTCGAATAGCGTTGTGACTGA-GGTAACTGCGCATCGAATCTACTCAGCGGCGAGGCTATTCCAGTAAGGTTGTTTTGGGC----CAGCGACTTTA--TGACC |
| droSec1 | super_1:1974074-1974203 + | TAGCCACGCAG--AGGA---------CACACCTG-------AAATTACCCATCTCGAATAGCGTTGTGACTGA-GGTAACTGCGCATCGAATCTACTCAGCGGCGAGGCTATTCCAGTAAGGTTGTTTTGGGC----CAGCGACTTTA--TGACC |
| droYak2 | chr2L:16988548-16988677 + | TAGAAACGCAG--AGGA---------CACACCTA-------AAATTACCCATCTCGAATAGCGTTGTGACTGA-GGTAACTGCGCATCGTATCTACTCAGCGGCGAAGCTATTCCAGTAAGGTTATTTTGGGC----CAGCGACTTTA--CGACC |
| droEre2 | scaffold_4929:18305702-18305825 - | TAGATACG--------A---------GATACCTA-------AGATTACCCATCTCGAATAGCGTTGTGACTGA-GGTAACTGCGCATCGAATCTACTCAGCGGCAAGGCTATTCCAGTAAGGTTATTTTGGGC----CAGCGACTCAA--CGACC |
| droAna3 | scaffold_13266:1670175-1670300 - | TGGTTTC-CTG--AGGA---------TACGTCCA-------AAATGACTCATCTCGAATAGCGTTGTGACTGATGGTATTTAAATAT------AAATCCGAGACAATGCTATTCTTGTAAAGTCATCTTGACC----AAGCAACTTTTTCTAATC |
| dp4 | chr3:2308599-2308727 + | TATCTCCTGAATCGGAG---------TACGACTG-------AAACAACCCGTCTCGAATAGCGTTGTGACTGAT-GA--CTGCATTTGTAATTTTCTCAGCGGCAGTGCTATTCCAGTTGGGTTGTTTGCGGCCAAACAACAACTATA------- |
| droPer1 | super_2:2480065-2480185 + | GAATCG--------GAG---------TACGACTG-------AAACAACCCGTCTCGAATAGCGTTGTGACTGAT-GA--CTGCATTTGTAATTTTCTCAGCGGCAGTGCTATTCCAGTTGGGTTGTTTGCGGCCAAACAACAACTATA------- |
| droWil1 | scaffold_180700:3161017-3161152 - | TTACTACACAA--AGTGAACAGTTC-GATGACTT-------AAATGCCCAATCTCGAATAGCGTTGTAACTGAT-GA--TTGT-GATATGAGAGAGTCAGCCACTATGCTATACGAGGTGGGGCATTTGTGGGT---AAACAACCAAA--TTACG |
| droVir3 | scaffold_12875:9403376-9403510 + | AAACTATAAAG--TG--TTTTAATGGTTCACTTAAATGGCTAATTGTCTTCTCTCGAATAGCATTATGGCTGAT-GACAAT---TCTGGAATAACATCAGTTGAAATGCCATTCGGGTTAAGGCACTTGAGAAATGGCAGCGA------------ |
| droMoj3 | scaffold_6496:17707880-17708014 - | TGTTGGCGAAC------TTTCAATGGCTCATTTAACTGGCTAATTGCCTTCTCTCGAATTGCATTATGGCCGAT-GACACAATTC-TGCAAAATCATCAGTTGGAATGCCATTCGGGTCAAGGCATTTACGAATCGCCAACGA------------ |
| droGri2 | scaffold_15245:2671305-2671410 + | TTGAATGGCT-------------------------------TATTGCCTTTTCTCGAATTGCATTGTGACTGAT-GAAAAT----TTAGAAATTCATCAGTTAAAATGCCATTCGAGTTAAGGCACTTATGCA-----------TTAA--CAACC |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chr2R:4332472-4332601 + | TAGCCACGCAG--AGGA---------CACACCTG-------AAATTACCCATCTCGAATAGCGTTGTGACTGA-GGTAACTGCGCATCGAATCTACTCAGCGGCGAGGCTATTCAAGTAAGGTTATTTTGGGC----CAGCGACTTTA--CGACC |
| droSim1 | chr2R:2956848-2956977 + | TAGCCACGCAG--AGGA---------CACACCTG-------AAATTACCCATCTCGAATAGCGTTGTGACTGA-GGTAACTGCGCATCGAATCTACTCAGCGGCGAGGCTATTCCAGTAAGGTTGTTTTGGGC----CAGCGACTTTA--TGACC |
| droSec1 | super_1:1974074-1974203 + | TAGCCACGCAG--AGGA---------CACACCTG-------AAATTACCCATCTCGAATAGCGTTGTGACTGA-GGTAACTGCGCATCGAATCTACTCAGCGGCGAGGCTATTCCAGTAAGGTTGTTTTGGGC----CAGCGACTTTA--TGACC |
| droYak2 | chr2L:16988548-16988677 + | TAGAAACGCAG--AGGA---------CACACCTA-------AAATTACCCATCTCGAATAGCGTTGTGACTGA-GGTAACTGCGCATCGTATCTACTCAGCGGCGAAGCTATTCCAGTAAGGTTATTTTGGGC----CAGCGACTTTA--CGACC |
| droEre2 | scaffold_4929:18305702-18305825 - | TAGATACG--------A---------GATACCTA-------AGATTACCCATCTCGAATAGCGTTGTGACTGA-GGTAACTGCGCATCGAATCTACTCAGCGGCAAGGCTATTCCAGTAAGGTTATTTTGGGC----CAGCGACTCAA--CGACC |
| droAna3 | scaffold_13266:1670175-1670300 - | TGGTTTC-CTG--AGGA---------TACGTCCA-------AAATGACTCATCTCGAATAGCGTTGTGACTGATGGTATTTAAATAT------AAATCCGAGACAATGCTATTCTTGTAAAGTCATCTTGACC----AAGCAACTTTTTCTAATC |
| dp4 | chr3:2308599-2308727 + | TATCTCCTGAATCGGAG---------TACGACTG-------AAACAACCCGTCTCGAATAGCGTTGTGACTGAT-GA--CTGCATTTGTAATTTTCTCAGCGGCAGTGCTATTCCAGTTGGGTTGTTTGCGGCCAAACAACAACTATA------- |
| droPer1 | super_2:2480065-2480185 + | GAATCG--------GAG---------TACGACTG-------AAACAACCCGTCTCGAATAGCGTTGTGACTGAT-GA--CTGCATTTGTAATTTTCTCAGCGGCAGTGCTATTCCAGTTGGGTTGTTTGCGGCCAAACAACAACTATA------- |
| droWil1 | scaffold_180700:3161017-3161152 - | TTACTACACAA--AGTGAACAGTTC-GATGACTT-------AAATGCCCAATCTCGAATAGCGTTGTAACTGAT-GA--TTGT-GATATGAGAGAGTCAGCCACTATGCTATACGAGGTGGGGCATTTGTGGGT---AAACAACCAAA--TTACG |
| droVir3 | scaffold_12875:9403376-9403510 + | AAACTATAAAG--TG--TTTTAATGGTTCACTTAAATGGCTAATTGTCTTCTCTCGAATAGCATTATGGCTGAT-GACAAT---TCTGGAATAACATCAGTTGAAATGCCATTCGGGTTAAGGCACTTGAGAAATGGCAGCGA------------ |
| droMoj3 | scaffold_6496:17707880-17708014 - | TGTTGGCGAAC------TTTCAATGGCTCATTTAACTGGCTAATTGCCTTCTCTCGAATTGCATTATGGCCGAT-GACACAATTC-TGCAAAATCATCAGTTGGAATGCCATTCGGGTCAAGGCATTTACGAATCGCCAACGA------------ |
| droGri2 | scaffold_15245:2671305-2671410 + | TTGAATGGCT-------------------------------TATTGCCTTTTCTCGAATTGCATTGTGACTGAT-GAAAAT----TTAGAAATTCATCAGTTAAAATGCCATTCGAGTTAAGGCACTTATGCA-----------TTAA--CAACC |
| Species | Read pileup | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
| dG=-26.4, p-value=0.009901 | dG=-26.3, p-value=0.009901 | dG=-25.6, p-value=0.009901 | dG=-25.5, p-value=0.009901 | dG=-25.5, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
| dG=-26.4, p-value=0.009901 | dG=-26.3, p-value=0.009901 | dG=-25.6, p-value=0.009901 | dG=-25.5, p-value=0.009901 | dG=-25.5, p-value=0.009901 |
|---|---|---|---|---|
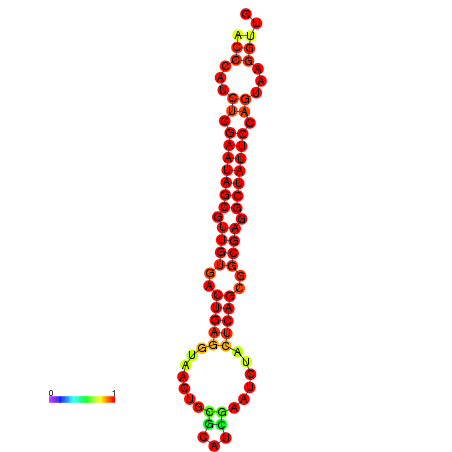 |
 |
 |
 |
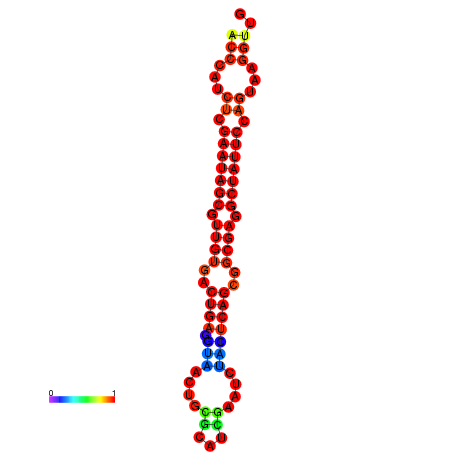 |
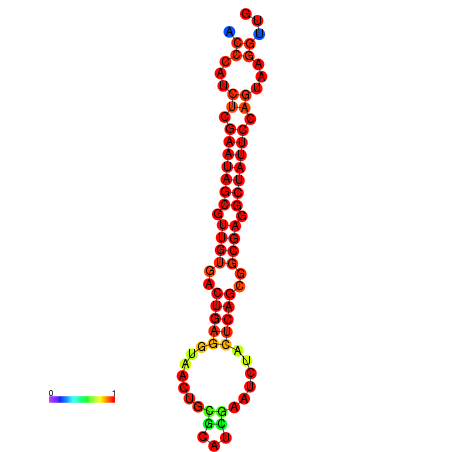
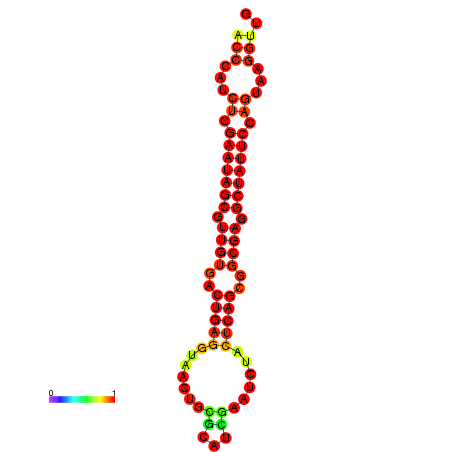
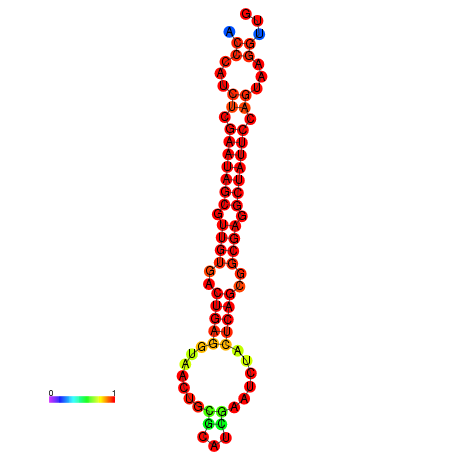
| dG=-26.4, p-value=0.009901 | dG=-26.3, p-value=0.009901 | dG=-25.6, p-value=0.009901 | dG=-25.5, p-value=0.009901 | dG=-25.5, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
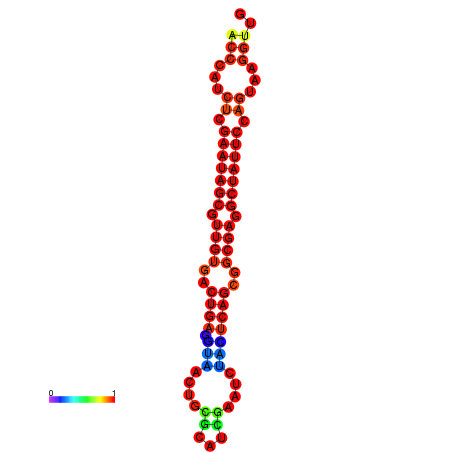 |
| dG=-26.2, p-value=0.009901 | dG=-26.2, p-value=0.009901 | dG=-26.2, p-value=0.009901 | dG=-25.4, p-value=0.009901 | dG=-25.4, p-value=0.009901 |
|---|---|---|---|---|
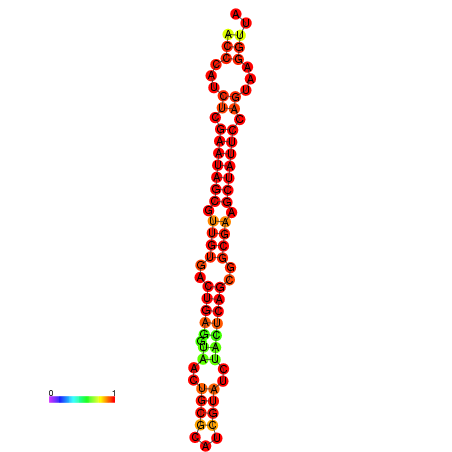 |
 |
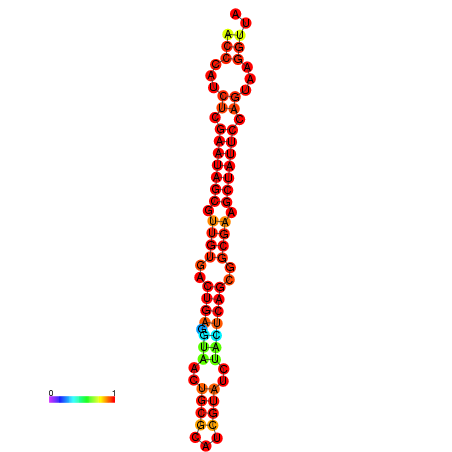 |
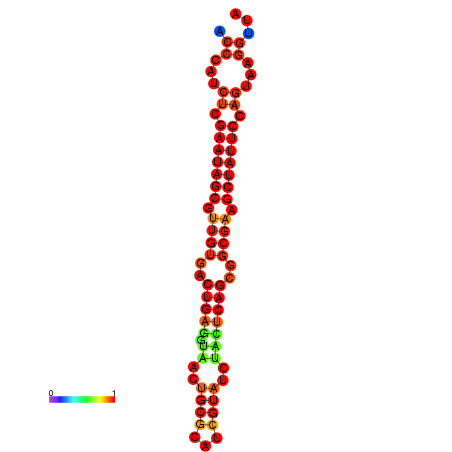 |
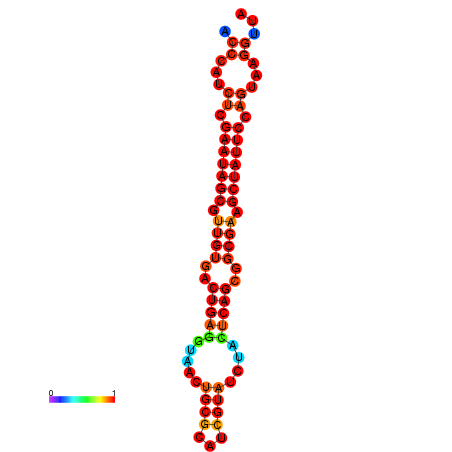 |
| dG=-26.7, p-value=0.009901 | dG=-26.6, p-value=0.009901 | dG=-25.9, p-value=0.009901 | dG=-25.8, p-value=0.009901 | dG=-25.8, p-value=0.009901 |
|---|---|---|---|---|
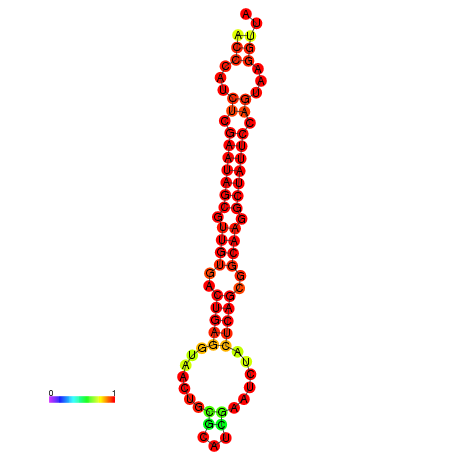 |
 |
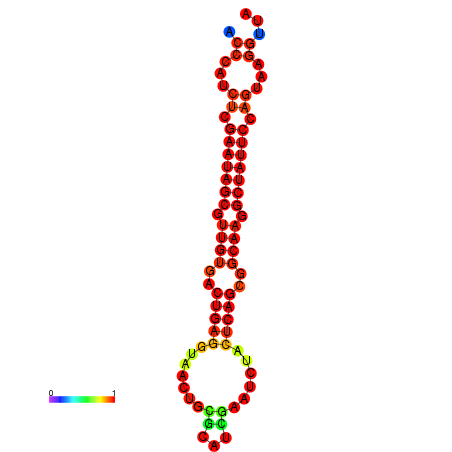 |
 |
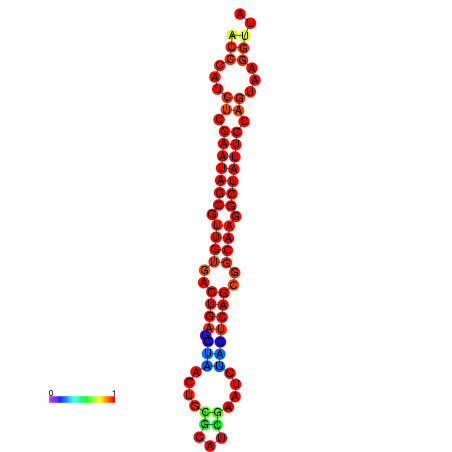 |
| dG=-17.4, p-value=0.009901 | dG=-17.3, p-value=0.009901 | dG=-17.2, p-value=0.009901 | dG=-17.0, p-value=0.009901 | dG=-16.9, p-value=0.009901 |
|---|---|---|---|---|
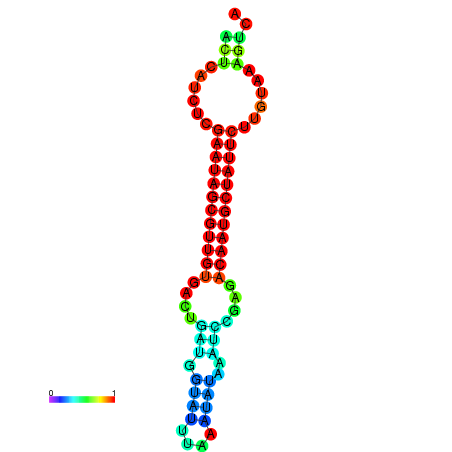 |
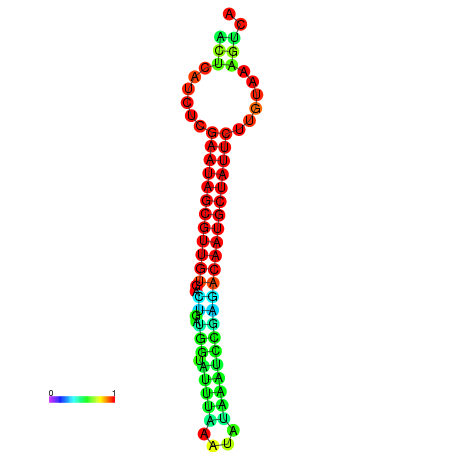 |
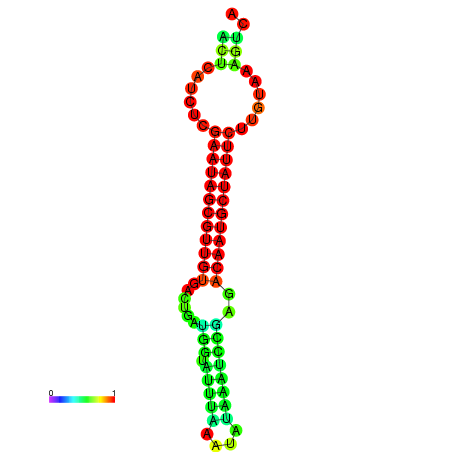 |
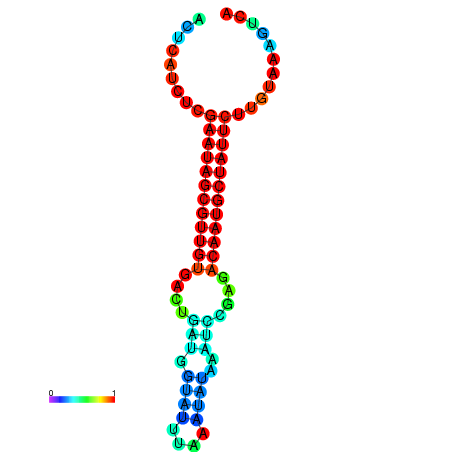 |
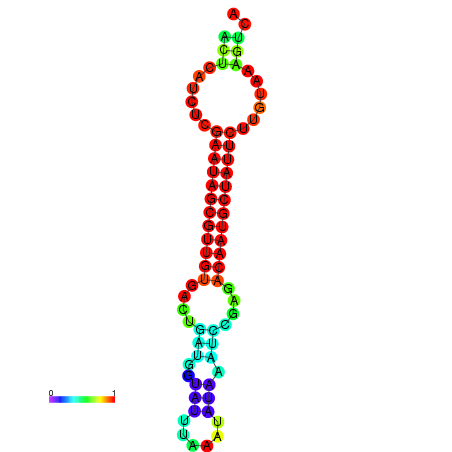 |
| dG=-27.3, p-value=0.009901 | dG=-27.3, p-value=0.009901 | dG=-27.1, p-value=0.009901 | dG=-26.7, p-value=0.009901 | dG=-26.7, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
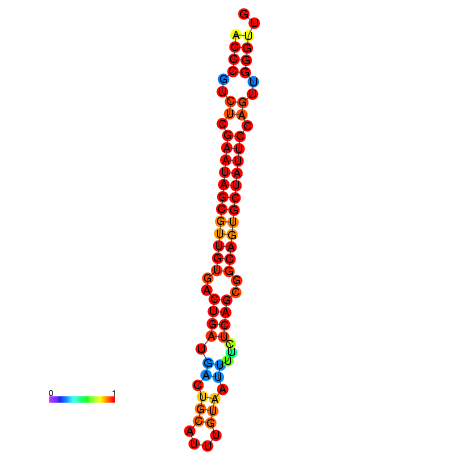 |
 |
| dG=-27.3, p-value=0.009901 | dG=-27.3, p-value=0.009901 | dG=-27.1, p-value=0.009901 | dG=-26.7, p-value=0.009901 | dG=-26.7, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
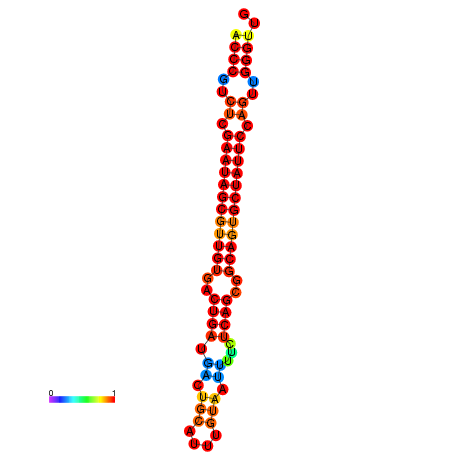 |
 |
| dG=-28.6, p-value=0.009901 | dG=-28.6, p-value=0.009901 | dG=-28.2, p-value=0.009901 |
|---|---|---|
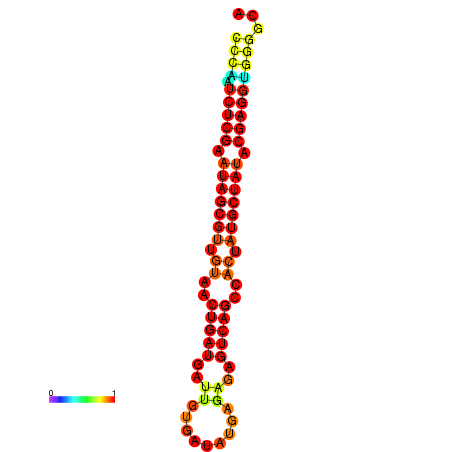 |
 |
 |
| dG=-24.8, p-value=0.009901 | dG=-24.7, p-value=0.009901 | dG=-24.2, p-value=0.009901 | dG=-24.1, p-value=0.009901 | dG=-23.9, p-value=0.009901 |
|---|---|---|---|---|
 |
 |
 |
 |
 |
| dG=-24.6, p-value=0.009901 | dG=-24.2, p-value=0.009901 | dG=-23.8, p-value=0.009901 | dG=-23.8, p-value=0.009901 |
|---|---|---|---|
 |
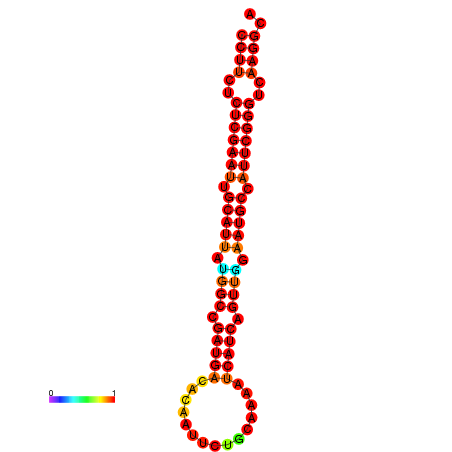 |
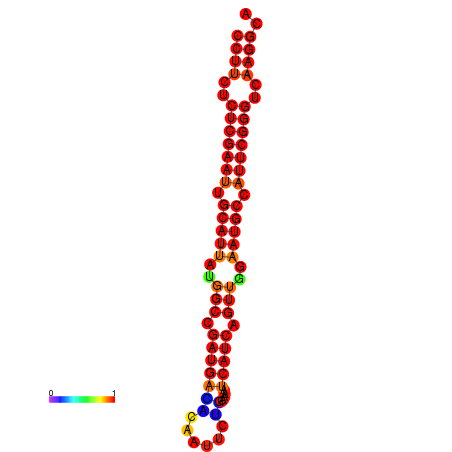 |
 |
| dG=-33.9, p-value=0.009901 | dG=-33.1, p-value=0.009901 | dG=-33.1, p-value=0.009901 | dG=-33.0, p-value=0.009901 |
|---|---|---|---|
 |
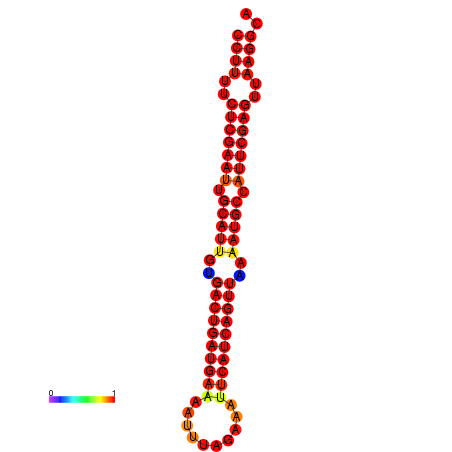 |
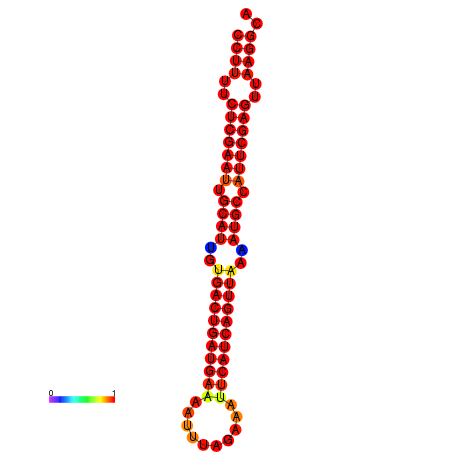 |
 |
Generated: 03/07/2013 at 07:20 PM