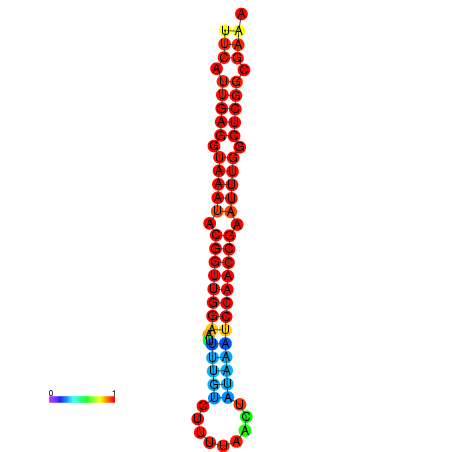
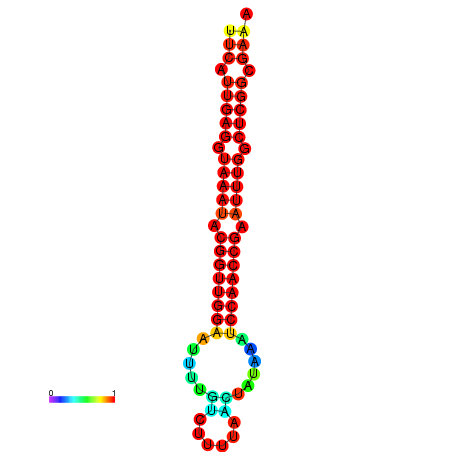
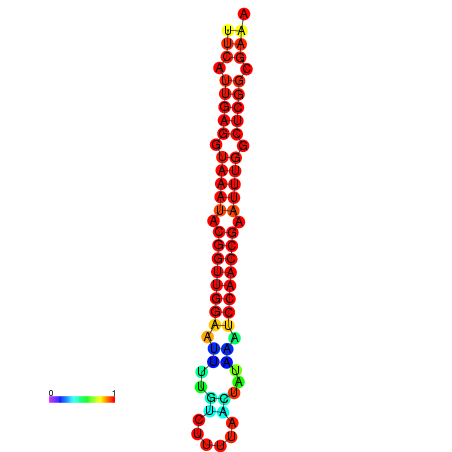
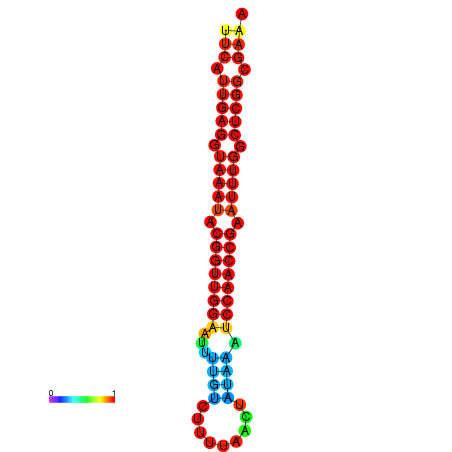
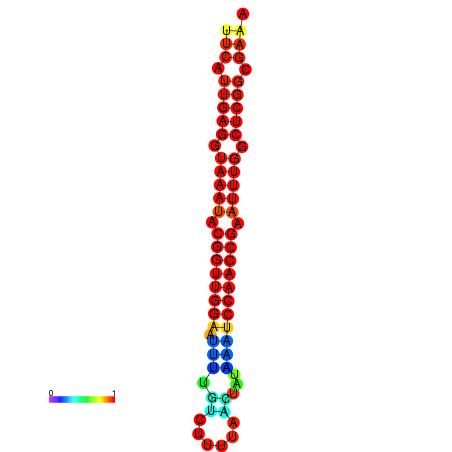
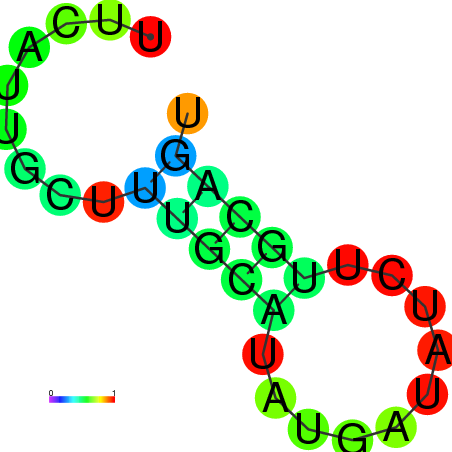
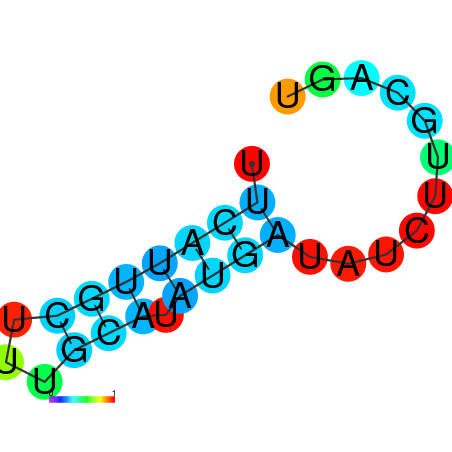
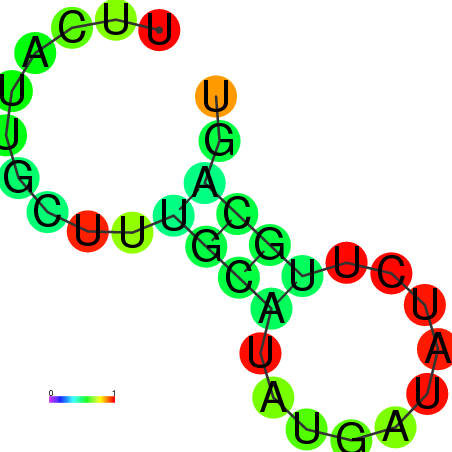
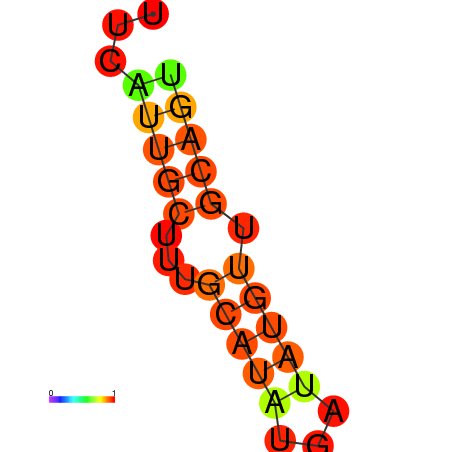
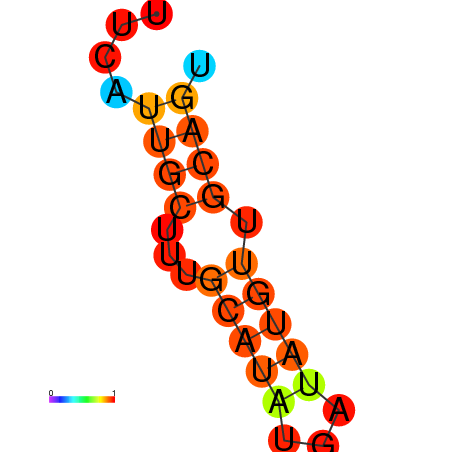
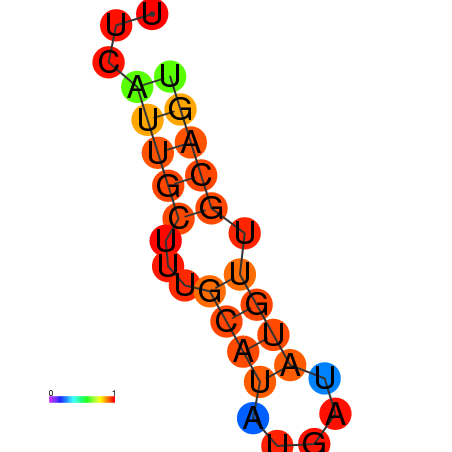
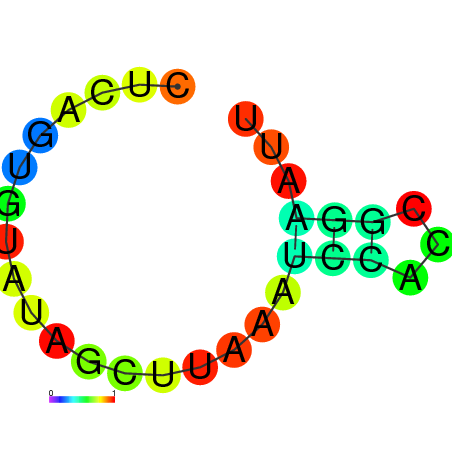
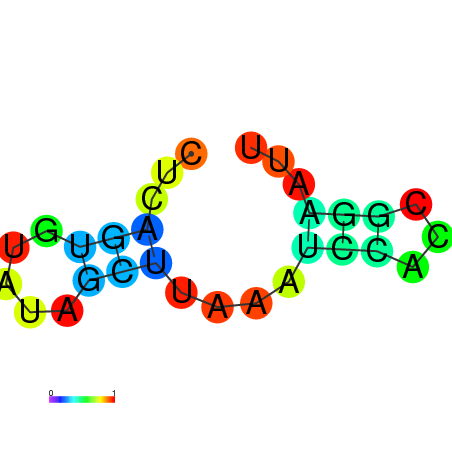
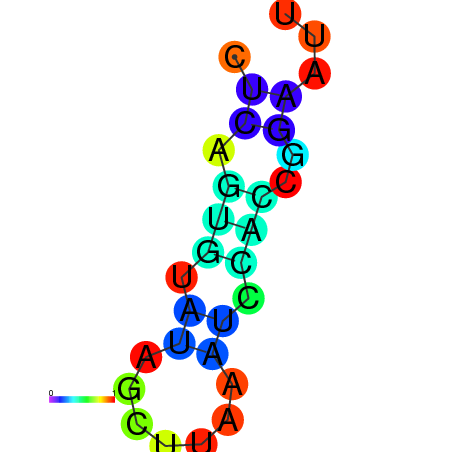
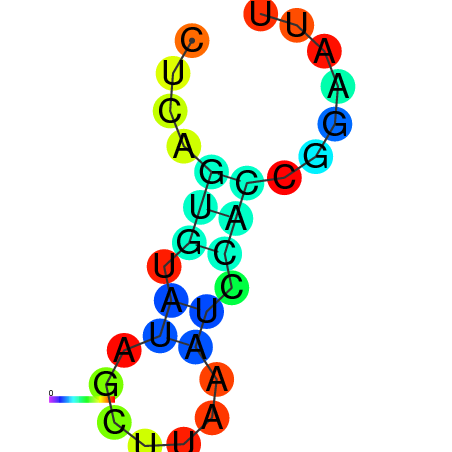
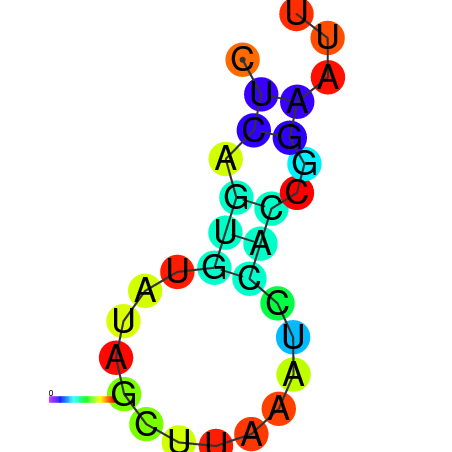
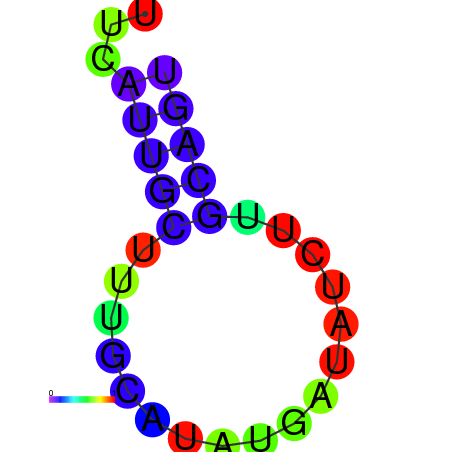| dm3 |
chrX:4262664-4262783 - |
TGGCTTCTATTGCGTAGAAACAAATTTCATTGAGGTAAATACGGTTGGAATTTTGTCTTTTAACTATAAATCCAACCGAATTTGGCTCGGCGAAATTTTTCAGTTTACTGGCGTCGAAAA |
| droSim1 |
chrX:3277428-3277474 - |
TGGCTTCTACTCTCT--------ACTTCATTGCTTTGCATATGAT---------------------------ATCTTGCAGT-------------------------------------- |
| droSec1 |
super_4:2462188-2462234 + |
TGGCTTCTACTATCT--------ACTTCATTGCTTTGCATATGAT---------------------------ATGTTGCAGT-------------------------------------- |
| droYak2 |
Unknown |
------------------------------------------------------------------------------------------------------------------------ |
| droEre2 |
scaffold_4690:1633226-1633277 - |
C---TTGTAA-ATGTACAAACAAATCTCAGTGTATAGCTTAAATC---------------------------CACCGGAATT-------------------------------------- |
| droAna3 |
Unknown |
------------------------------------------------------------------------------------------------------------------------ |
| dp4 |
Unknown |
------------------------------------------------------------------------------------------------------------------------ |
| droPer1 |
Unknown |
------------------------------------------------------------------------------------------------------------------------ |
| droWil1 |
Unknown |
------------------------------------------------------------------------------------------------------------------------ |
| droVir3 |
Unknown |
------------------------------------------------------------------------------------------------------------------------ |
| droMoj3 |
scaffold_6359:3791288-3791288 - |
T----------------------------------------------------------------------------------------------------------------------- |
| droGri2 |
Unknown |
------------------------------------------------------------------------------------------------------------------------ |