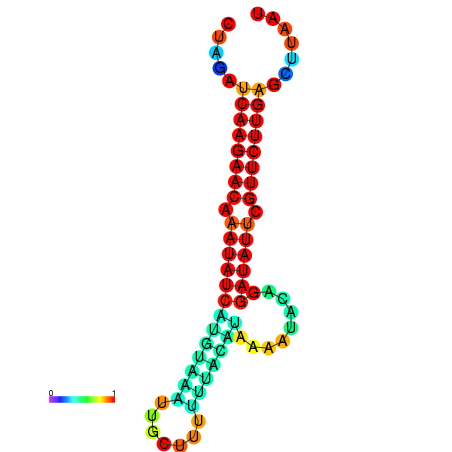
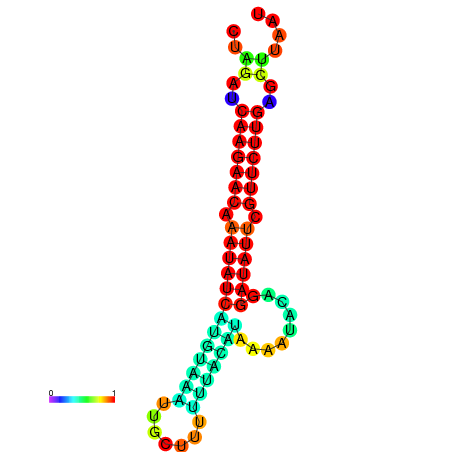
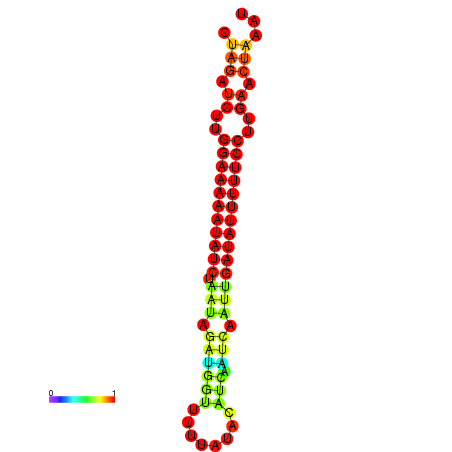
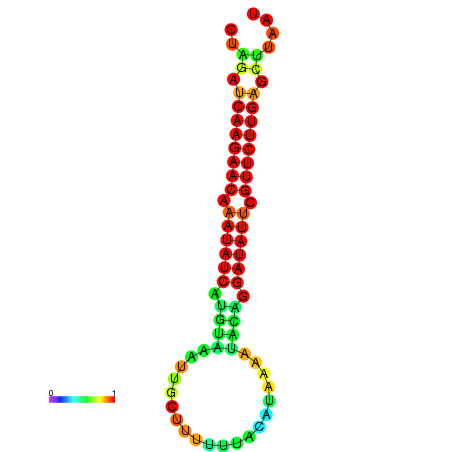| dm3 |
chrX:4259824-4259946 - |
TATCAAGTTAAAAAA------CGAAATCATGTTAGATCCTGGACAAATATGAAGTAAATTGTTTTTA----TGCAT--C-----------AA-TTACTTGATATT-CATCCTTG-AACTAAATG----------GTTT-TAGAGCTGCAAGA----AAAATTGT |
| droSim1 |
chrX:3274541-3274667 - |
TATCAAGTTAAAAAA------CGAAATCATGCTAGATCATTAACAAGTATCATGTAAATTGAATTTTTGTATATAT--C-----------AA-TTACATGTTATT-CGTTCTTG-AACTTAATG----------GTTT-TAGAGCTGCAAGA----AAAATTGT |
| droSec1 |
super_4:2464986-2465108 + |
TATGAAGTTAAAAAA------CGAAATCATGCTAGATCAAGAACAAATATCATGTAAATTGCTTTTT----TACAT--A-----------AA-ATACAGGATATT-CGTTCTTG-AGCTTAATG----------GTTA-TAGAGCTGCAAGA----AAAATTGT |
| droYak2 |
chrX:4178368-4178492 + |
TATCAAGTTCAGAAA----TATAAAATCATGCTAGATCTTGGAAAAATATCTAATAGATGGTTTTTA----TACAT--C-----------AA-TCAATTGATATT-TTTCCTTG-AACTAAATG----------GGTT-TAGTGCTGCAGCA----AAAATGGT |
| droEre2 |
scaffold_4690:1630508-1630651 - |
TATCAAGTTAAAAAA----TATAAAAGCAGTCCAGATCATGGAAAAATATCAAATAGATTGTTTTAT----TTCAGTCCAAACAATACATAA-TCACGCGAGTCA-TAGTATCGAAATTTAGCA----------ATTTATGAAGTTAAAAAAATATAAAGCCAT |
| droAna3 |
scaffold_13335:212413-212477 + |
TTTGTGGATC--------------------------------------------------------------------------------GA-TTTCCAGTAATT-CTGCATCA-AAATCCGG-ACAAAAAATATTTT-T----------TG----AAAATTTT |
| dp4 |
Unknown |
-------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droPer1 |
Unknown |
-------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droWil1 |
Unknown |
-------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droVir3 |
Unknown |
-------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droMoj3 |
scaffold_6496:5428540-5428655 - |
TTG-TTGTTGGCTGTTAACAACTTGTTCGATTTGGCCTCTGGGTAGACTCTATATA-----------------TGT--A-----------AATATATAT-ATATCTAATCTCTA-ATATCTCAC----------GTTT-TACACCTGCAAAA----AAAACTTC |
| droGri2 |
Unknown |
-------------------------------------------------------------------------------------------------------------------------------------------------------------------- |