| Legend: | mature | star | mismatch |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chrX:174190-174317 - | GCGTG-------------------TGAAATACGAAGTTGATTGTATGTCAGTTTTTCATTTGGCCTGGCTAGCTTACTCCT--TTTTAAATATTGCTAGCTGCCTTGTGAAGGGCTTACGTGTAATTGCAGTTCGAC-----------TCCCGAATA---ATA |
| droSim1 | chrX:118257-118384 - | GCGTG-------------------TGAAATACGAAGTTGATTGTATGTCAGTTTTTCATTTGGCGTGGCTAGCTCACTACT--TTTTTAATATTGCTAGCTGCCTTGTGAAGGGCTTACGTGTAATTGCAGTTCAAC-----------TGCCAAATA---ATA |
| droSec1 | super_10:47183-47310 - | GCGTG-------------------TGAAATACGAAGTTGATTGTATGTCAGTTTTTCATTTGGCGTGGCTAGCTCACTACT--TTTTTAATATTGCTAGCTGCCTTGTGAAGGGCTTACGTGTAATTGCAGTTCAAC-----------TGCCAAATA---ATA |
| droYak2 | chrX:117050-117182 - | GCGTG-------------------TGAGATGTGAAGTTGATTGTATGTCAGTTTTTCATTTGGCGTGCCTAGCTTAGCTTT--TTTTAAATATTGCTAGCTGCCTTGTGAAGGGCTTACGTGTAATTGCAGTTCGACTCGA------CTGCCAAATG---ATA |
| droEre2 | scaffold_4644:121637-121764 - | GCGTG-------------------TGAAATGTGAAGTTGATTGTATGTCAGTTTTTCATTTGGCGTGCCTAGCTTACCTCT--TTTTAAATATTGCTAGCTGCCTTGTGAAGGGCTTACGTGTAATTGTAGTTCGAC-----------TGCAAAATT---ATA |
| droAna3 | scaffold_13117:4815182-4815314 - | AAGTGATAAGAAATGTCC-----------TAGGAAGTAATTTGTATGTCAGCCTCTCATTTGGCCTGCCTAGCTGACTTT---AAATTAATATTGCTAGCTGCCTTGTGAAGGGCTTACGTGTGATTGCGATTCAAC-----------TTGC--AGA---ATA |
| dp4 | chrXL_group1e:4097625-4097758 - | CCGCC-------------------ACAAAAGAGAAACAAATTGAATGTGAGTCTCTCACATGGCTGGTCTAGCTGACTTG---TTTCTAATATTGCTAGCTGCCTTGTGAAGGGCTTACGTGTTTTTGTAGCCCAACGTAA-------AGCCGAAAATCGTAA |
| droPer1 | super_17:1039174-1039307 - | CCGCC-------------------ACAAAAGAGAAACAAATTGTATGTGAGTCTCTCACATGGCTGGTCTAGCTGACTTG---TTTCTAATACTGCTAGCTGCCTTGTGAAGGGCTTACGTGTTTTTGTAGCCCAACGTAA-------AGCCGAAAATCGTAA |
| droWil1 | scaffold_181096:8731771-8731891 + | ---------------------------------------ATTGTGTGTGAGTCTTTCACTGGGCGTGCTTAGCTAGAAATTTATTTACCATATTGCTAGCTGCCTTGTGAAGTGCTTACGTGTAATTGTTGAACAACAAAAAAAAATCAGAAAAAGC---ATA |
| droVir3 | scaffold_10277:1011-1148 - | GGGCTATAAAATTTGGACCAAATGTGA------GAGCGCATTGCATGTGAGTCCCTCATTTGGCAGGTTTAGCTGCTTTT---AATACGATATTGCTAGCTGCCTTGTGAAGGGCTTACGTGTGATTGTTAAACAAC-----------T--TAAACA---ACA |
| droMoj3 | scaffold_6473:11075547-11075659 + | TTATT-------------------T---ATATGAACCGCATTGCATGTGAGTCCCTCATTTGGTGCGCTTAGCTGTTTTTC--AATTTGATATTGCTAGCTGCCTTGTGAAGGGCTTACGTGTGTTTGTAATACAAC-------------------------- |
| droGri2 | scaffold_14853:6339037-6339158 + | GATTA-------------------TGAG------AGCGCATTACATGTGAGTCCCTCATTTGGCGGACTTAGCTGCTTTTC--AATTTGATATTGCTAGCTGCCTTGTGAAGGGCTTACGTGTGATTGTAAATCAAC-----------TGAGGAAAA---ATA |
| Species | Coordinate | Alignment |
|---|---|---|
| dm3 | chrX:174190-174317 - | GCGTG-------------------TGAAATACGAAGTTGATTGTATGTCAGTTTTTCATTTGGCCTGGCTAGCTTACTCCT--TTTTAAATATTGCTAGCTGCCTTGTGAAGGGCTTACGTGTAATTGCAGTTCGAC-----------TCCCGAATA---ATA |
| droSim1 | chrX:118257-118384 - | GCGTG-------------------TGAAATACGAAGTTGATTGTATGTCAGTTTTTCATTTGGCGTGGCTAGCTCACTACT--TTTTTAATATTGCTAGCTGCCTTGTGAAGGGCTTACGTGTAATTGCAGTTCAAC-----------TGCCAAATA---ATA |
| droSec1 | super_10:47183-47310 - | GCGTG-------------------TGAAATACGAAGTTGATTGTATGTCAGTTTTTCATTTGGCGTGGCTAGCTCACTACT--TTTTTAATATTGCTAGCTGCCTTGTGAAGGGCTTACGTGTAATTGCAGTTCAAC-----------TGCCAAATA---ATA |
| droYak2 | chrX:117050-117182 - | GCGTG-------------------TGAGATGTGAAGTTGATTGTATGTCAGTTTTTCATTTGGCGTGCCTAGCTTAGCTTT--TTTTAAATATTGCTAGCTGCCTTGTGAAGGGCTTACGTGTAATTGCAGTTCGACTCGA------CTGCCAAATG---ATA |
| droEre2 | scaffold_4644:121637-121764 - | GCGTG-------------------TGAAATGTGAAGTTGATTGTATGTCAGTTTTTCATTTGGCGTGCCTAGCTTACCTCT--TTTTAAATATTGCTAGCTGCCTTGTGAAGGGCTTACGTGTAATTGTAGTTCGAC-----------TGCAAAATT---ATA |
| droAna3 | scaffold_13117:4815182-4815314 - | AAGTGATAAGAAATGTCC-----------TAGGAAGTAATTTGTATGTCAGCCTCTCATTTGGCCTGCCTAGCTGACTTT---AAATTAATATTGCTAGCTGCCTTGTGAAGGGCTTACGTGTGATTGCGATTCAAC-----------TTGC--AGA---ATA |
| dp4 | chrXL_group1e:4097625-4097758 - | CCGCC-------------------ACAAAAGAGAAACAAATTGAATGTGAGTCTCTCACATGGCTGGTCTAGCTGACTTG---TTTCTAATATTGCTAGCTGCCTTGTGAAGGGCTTACGTGTTTTTGTAGCCCAACGTAA-------AGCCGAAAATCGTAA |
| droPer1 | super_17:1039174-1039307 - | CCGCC-------------------ACAAAAGAGAAACAAATTGTATGTGAGTCTCTCACATGGCTGGTCTAGCTGACTTG---TTTCTAATACTGCTAGCTGCCTTGTGAAGGGCTTACGTGTTTTTGTAGCCCAACGTAA-------AGCCGAAAATCGTAA |
| droWil1 | scaffold_181096:8731771-8731891 + | ---------------------------------------ATTGTGTGTGAGTCTTTCACTGGGCGTGCTTAGCTAGAAATTTATTTACCATATTGCTAGCTGCCTTGTGAAGTGCTTACGTGTAATTGTTGAACAACAAAAAAAAATCAGAAAAAGC---ATA |
| droVir3 | scaffold_10277:1011-1148 - | GGGCTATAAAATTTGGACCAAATGTGA------GAGCGCATTGCATGTGAGTCCCTCATTTGGCAGGTTTAGCTGCTTTT---AATACGATATTGCTAGCTGCCTTGTGAAGGGCTTACGTGTGATTGTTAAACAAC-----------T--TAAACA---ACA |
| droMoj3 | scaffold_6473:11075547-11075659 + | TTATT-------------------T---ATATGAACCGCATTGCATGTGAGTCCCTCATTTGGTGCGCTTAGCTGTTTTTC--AATTTGATATTGCTAGCTGCCTTGTGAAGGGCTTACGTGTGTTTGTAATACAAC-------------------------- |
| droGri2 | scaffold_14853:6339037-6339158 + | GATTA-------------------TGAG------AGCGCATTACATGTGAGTCCCTCATTTGGCGGACTTAGCTGCTTTTC--AATTTGATATTGCTAGCTGCCTTGTGAAGGGCTTACGTGTGATTGTAAATCAAC-----------TGAGGAAAA---ATA |
| Species | Read pileup | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droSim1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp4 |
|
| dG=-27.9, p-value=0.009901 |
|---|
 |
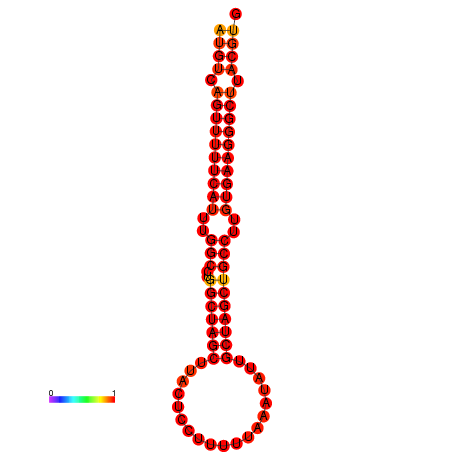
| dG=-27.9, p-value=0.009901 | dG=-27.2, p-value=0.009901 |
|---|---|
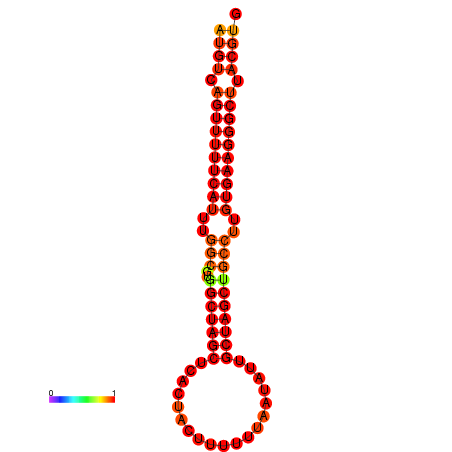 |
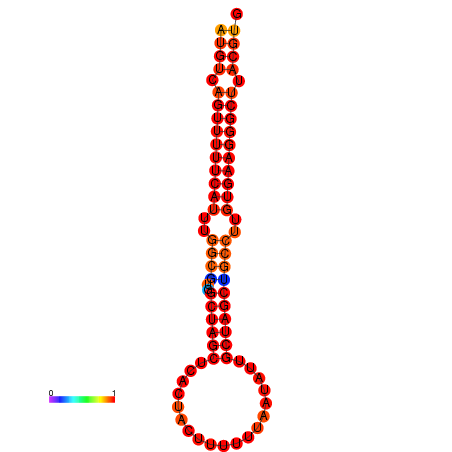 |
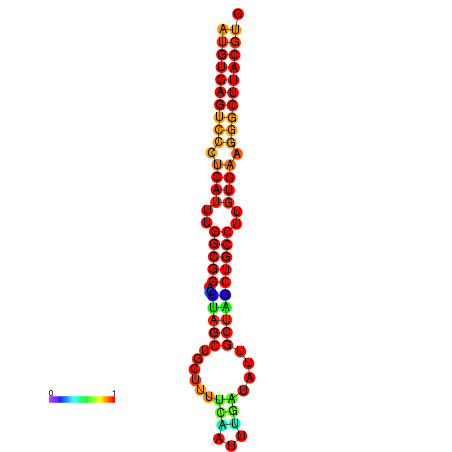
| dG=-27.9, p-value=0.009901 | dG=-27.2, p-value=0.009901 |
|---|---|
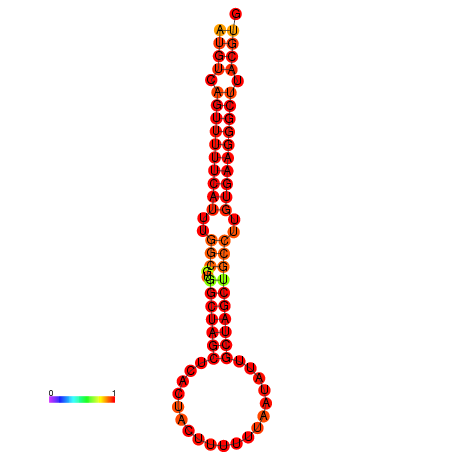 |
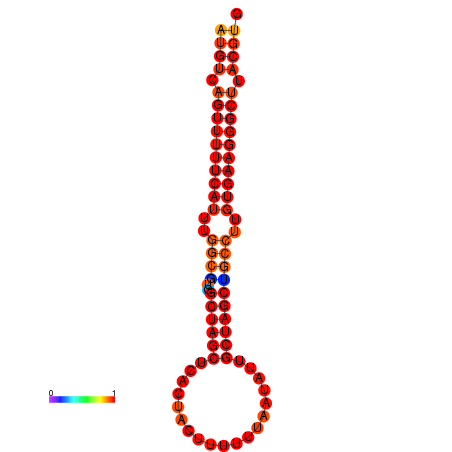 |
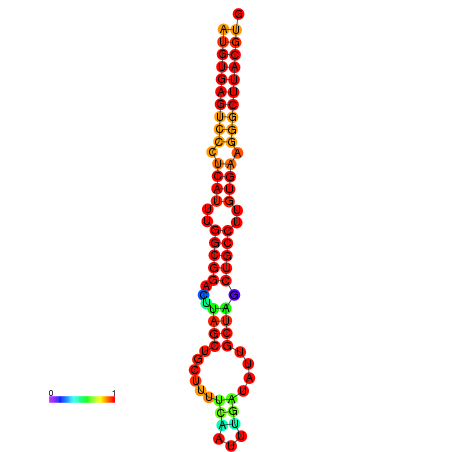
| dG=-25.0, p-value=0.009901 | dG=-24.1, p-value=0.009901 | dG=-24.1, p-value=0.009901 |
|---|---|---|
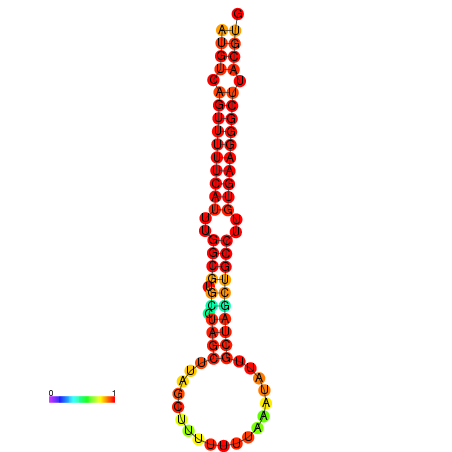 |
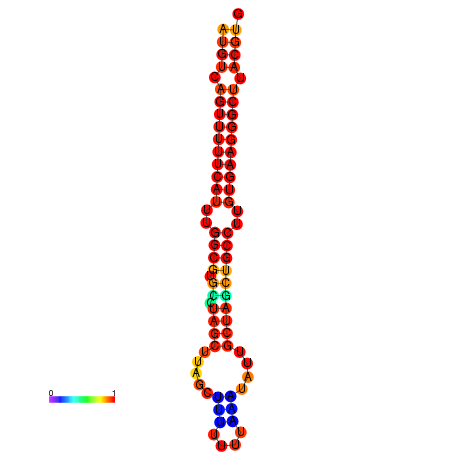 |
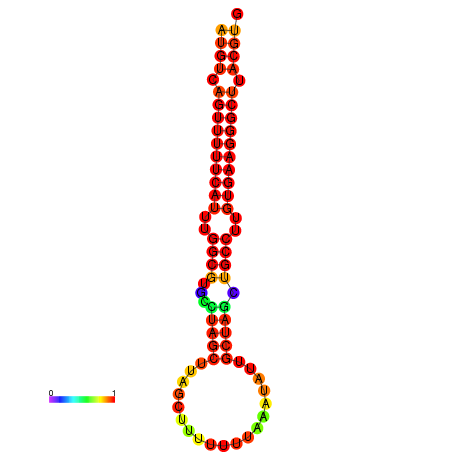 |
| dG=-25.0, p-value=0.009901 | dG=-24.1, p-value=0.009901 |
|---|---|
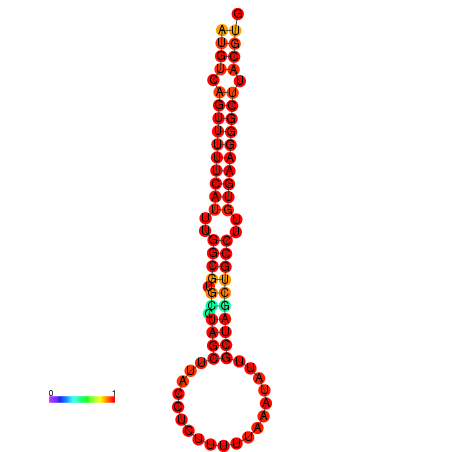 |
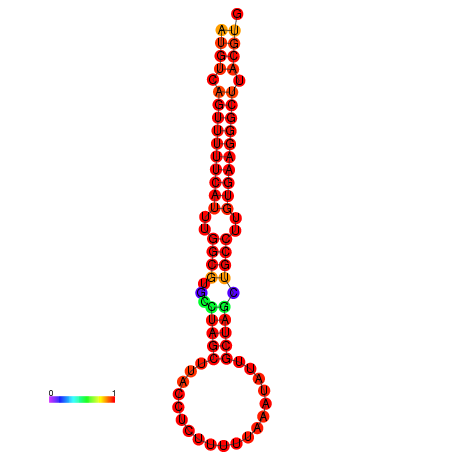 |
| dG=-25.8, p-value=0.009901 | dG=-24.8, p-value=0.009901 |
|---|---|
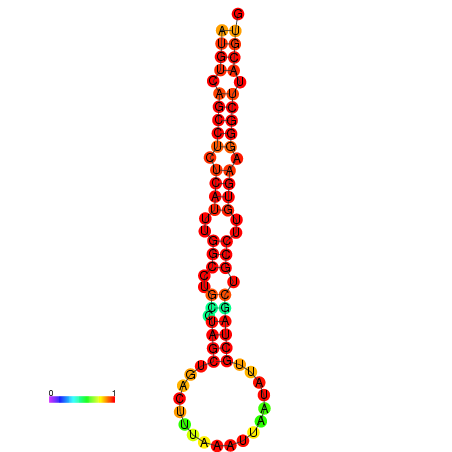 |
 |
| dG=-31.8, p-value=0.009901 | dG=-31.0, p-value=0.009901 | dG=-30.8, p-value=0.009901 |
|---|---|---|
 |
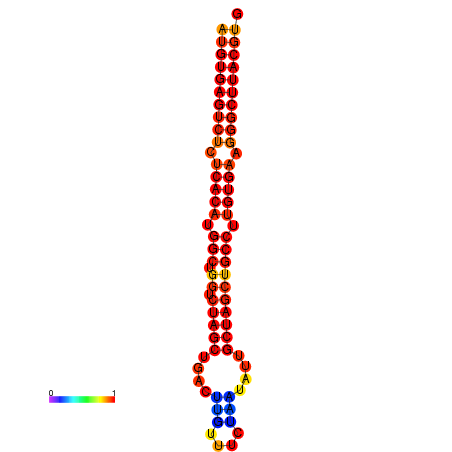 |
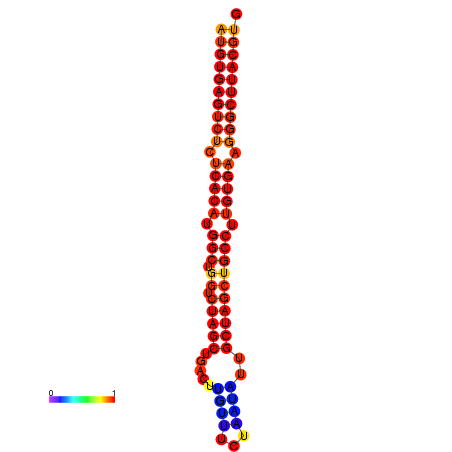 |
| dG=-31.8, p-value=0.009901 | dG=-31.0, p-value=0.009901 |
|---|---|
 |
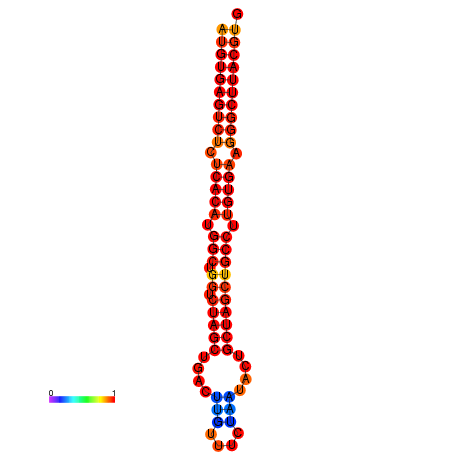 |
| dG=-27.8, p-value=0.009901 | dG=-27.8, p-value=0.009901 | dG=-27.2, p-value=0.009901 | dG=-27.2, p-value=0.009901 |
|---|---|---|---|
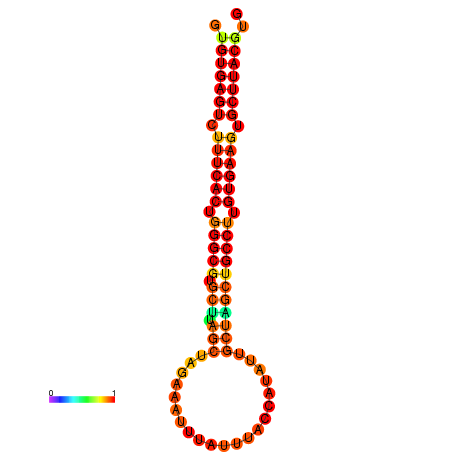 |
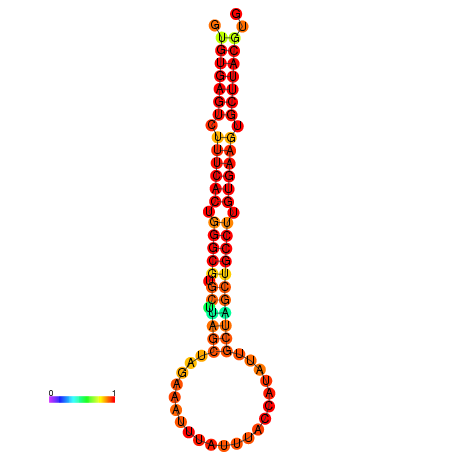 |
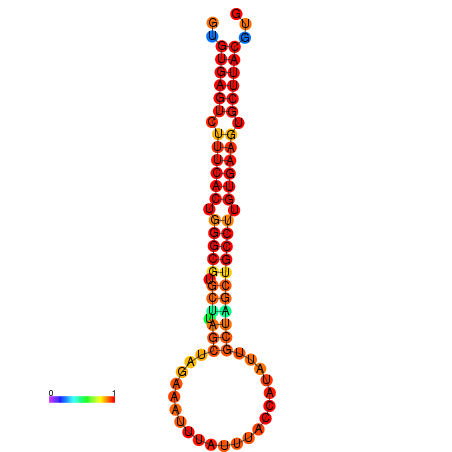 |
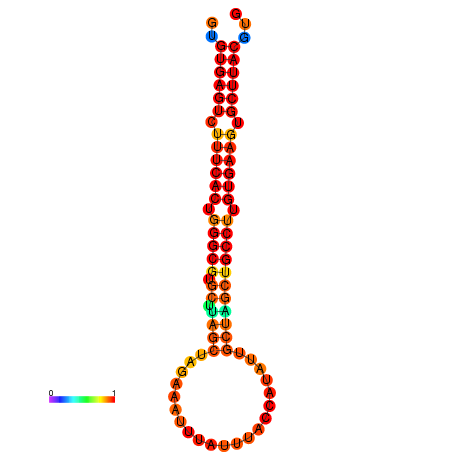 |
| dG=-30.5, p-value=0.009901 | dG=-30.2, p-value=0.009901 | dG=-29.7, p-value=0.009901 |
|---|---|---|
 |
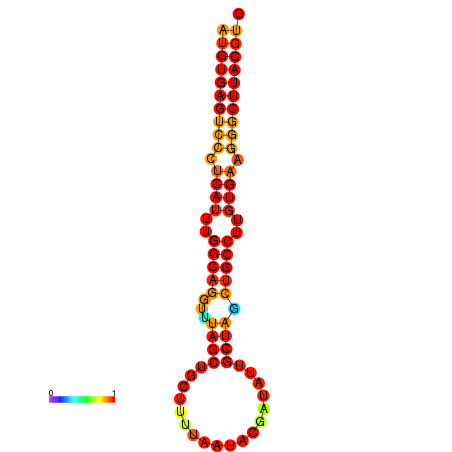 |
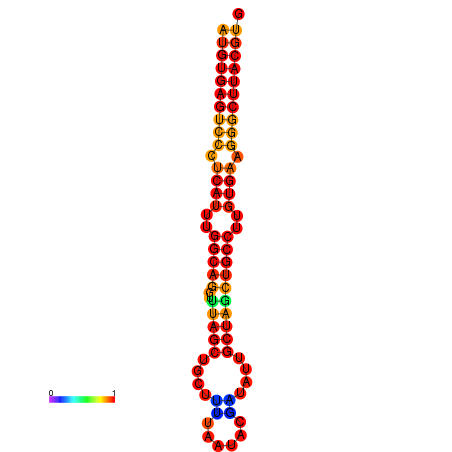 |
| dG=-28.2, p-value=0.009901 | dG=-28.2, p-value=0.009901 | dG=-28.0, p-value=0.009901 | dG=-28.0, p-value=0.009901 | dG=-27.8, p-value=0.009901 |
|---|---|---|---|---|
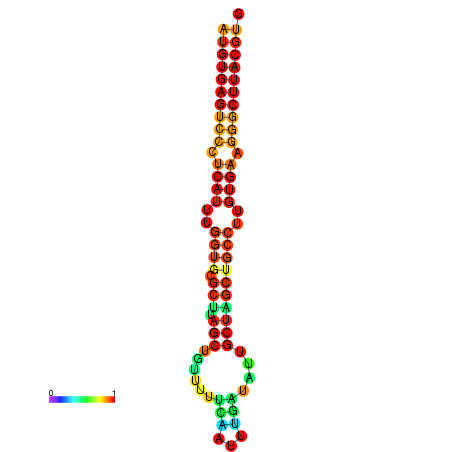 |
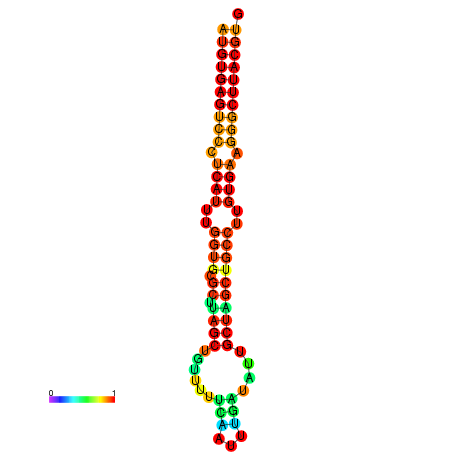 |
 |
 |
 |
| dG=-30.8, p-value=0.009901 | dG=-30.4, p-value=0.009901 | dG=-30.3, p-value=0.009901 | dG=-30.2, p-value=0.009901 | dG=-29.9, p-value=0.009901 |
|---|---|---|---|---|
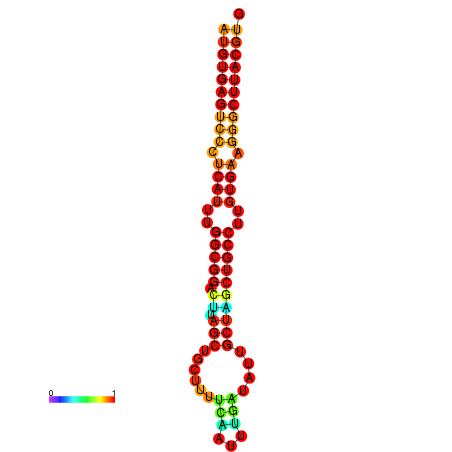 |
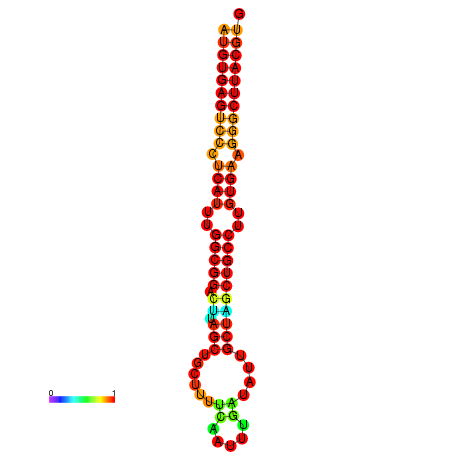 |
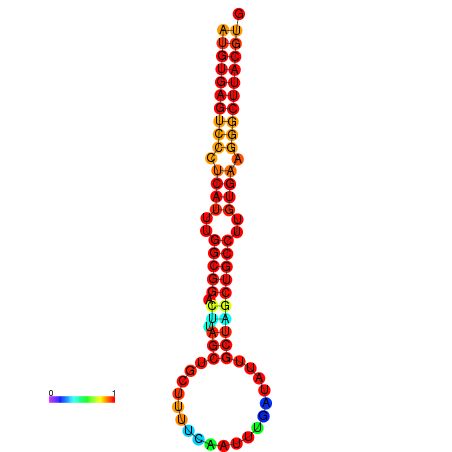 |
 |
 |
Generated: 03/07/2013 at 07:19 PM